Snapshot: Axon Guidance II Alex L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
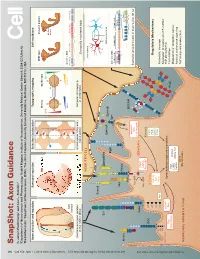
Snapshot: Axon Guidance Pasterkamp R
494 1 Cell Cell ??? SnapShot: Axon Guidance 153 SnapShot: XXXXXXXXXXXXXXXXXXXXXXXXXX 1 2 , ??MONTH?? ??DATE??, 200? ©200? Elsevier Inc. 200?©200? ElsevierInc. , ??MONTH?? ??DATE??, DOI R. Jeroen Pasterkamp and Alex L. Kolodkin , April11, 2013©2013Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.03.031 AUTHOR XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 1 AFFILIATIONDepartment of XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX Neuroscience and Pharmacology, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, 3584 CG Utrecht, The Netherlands; 2Department of Neuroscience, HHMI, The Johns Hopkins University School of Medicine, Baltimore, MD 21212, USA Axon attraction and repulsion Surround repulsion Selective fasciculation Topographic mapping Self-avoidance Wild-type Dscam1 mutant Sema3A A Retina SC/Tectum EphB EphrinB P D D Mutant T A neuron N P A V V Genomic DNA P EphA EphrinA Slit Exon 4 (12) Exon 6 (48) Exon 9 (33) Exon 17 (2) Netrin Commissural axon guidance Surround repulsion of peripheral Grasshopper CNS axon Retinotectal mapping at the CNS midline nerves in vertebrates fasciculation in vertebrates Drosophila mushroom body XXXXXXXXX NEURITE/CELL Isoneuronal Sema3 Slit Sema1/4-6 Heteroneuronal EphrinA EphrinB FasII Eph Genomic DNA Pcdh-α (14) Pcdh-β (22) Pcdh-γ (22) Variable Con Variable Con Nrp Plexin ** *** Netrin LAMELLIPODIA Con DSCAM ephexin Starburst amacrine cells in mammalian retina Ras-GTP Vav See online version for legend and references. α-chimaerin GEFs/GAPs Robo FARP Ras-GDP LARG RhoGEF Kinases DCC cc0 GTPases PKA cc1 FAK Regulatory Mechanisms See online versionfor??????. Cdc42 GSK3 cc2 Rac PI3K P1 Rho P2 cc3 Abl Proteolytic cleavage P3 Regulation of expression (TF, miRNA, FILOPODIA srGAP Cytoskeleton regulatory proteins multiple isoforms) Sos Trio Pcdh Cis inhibition DOCK180 PAK ROCK Modulation of receptors’ output LIMK Myosin-II Colin Forward and reverse signaling Actin Trafcking and endocytosis NEURONAL GROWTH CONE Microtubules SnapShot: Axon Guidance R. -

Regulation of the Mammalian Target of Rapamycin Complex 2 (Mtorc2)
Regulation of the Mammalian Target Of Rapamycin Complex 2 (mTORC2) Inauguraldissertation Zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Klaus-Dieter Molle aus Heilbronn, Deutschland Basel, 2006 Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät Auf Antrag von Prof. Michael N. Hall und Prof. Markus Affolter. Basel, den 21.11.2006 Prof. Hans-Peter Hauri Dekan Summary The growth controlling mammalian Target of Rapamycin (mTOR) is a conserved Ser/Thr kinase found in two structurally and functionally distinct complexes, mTORC1 and mTORC2. The tumor suppressor TSC1-TSC2 complex inhibits mTORC1 by acting on the small GTPase Rheb, but the role of TSC1-TSC2 and Rheb in the regulation of mTORC2 is unclear. Here we examined the role of TSC1-TSC2 in the regulation of mTORC2 in human embryonic kidney 293 cells. Induced knockdown of TSC1 and TSC2 (TSC1/2) stimulated mTORC2-dependent actin cytoskeleton organization and Paxillin phosphorylation. Furthermore, TSC1/2 siRNA increased mTORC2-dependent Ser473 phosphorylation of plasma membrane bound, myristoylated Akt/PKB. This suggests that loss of Akt/PKB Ser473 phosphorylation in TSC mutant cells, as reported previously, is due to inhibition of Akt/PKB localization rather than inhibition of mTORC2 activity. Amino acids and overexpression of Rheb failed to stimulate mTORC2 signaling. Thus, TSC1-TSC2 also inhibits mTORC2, but possibly independently of Rheb. Our results suggest that mTORC2 hyperactivation may contribute to the pathophysiology of diseases such as cancer and Tuberous Sclerosis Complex. i Acknowledgement During my PhD studies in the Biozentrum I received a lot of support from many people around me who I mention here to express my gratefulness. -

The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination
The Journal of Neuroscience, August 31, 2011 • 31(35):12579–12592 • 12579 Cellular/Molecular The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination Junji Yamauchi,1,3,5 Yuki Miyamoto,1 Hajime Hamasaki,1,3 Atsushi Sanbe,1 Shinji Kusakawa,1 Akane Nakamura,2 Hideki Tsumura,2 Masahiro Maeda,4 Noriko Nemoto,6 Katsumasa Kawahara,5 Tomohiro Torii,1 and Akito Tanoue1 1Department of Pharmacology and 2Laboratory Animal Resource Facility, National Research Institute for Child Health and Development, Setagaya, Tokyo 157-8535, Japan, 3Department of Biological Sciences, Tokyo Institute of Technology, Midori, Yokohama 226-8501, Japan, 4IBL, Ltd., Fujioka, Gumma 375-0005, Japan, and 5Department of Physiology and 6Bioimaging Research Center, Kitasato University School of Medicine, Sagamihara, Kanagawa 252-0374, Japan In development of the peripheral nervous system, Schwann cells proliferate, migrate, and ultimately differentiate to form myelin sheath. In all of the myelination stages, Schwann cells continuously undergo morphological changes; however, little is known about their underlying molecular mechanisms. We previously cloned the dock7 gene encoding the atypical Rho family guanine-nucleotide exchange factor (GEF) and reported the positive role of Dock7, the target Rho GTPases Rac/Cdc42, and the downstream c-Jun N-terminal kinase in Schwann cell migration (Yamauchi et al., 2008). We investigated the role of Dock7 in Schwann cell differentiation and myelination. Knockdown of Dock7 by the specific small interfering (si)RNA in primary Schwann cells promotes dibutyryl cAMP-induced morpholog- ical differentiation, indicating the negative role of Dock7 in Schwann cell differentiation. It also results in a shorter duration of activation of Rac/Cdc42 and JNK, which is the negative regulator of myelination, and the earlier activation of Rho and Rho-kinase, which is the positive regulator of myelination. -

Identification of Two Signaling Submodules Within the Crkii/ELMO
Cell Death and Differentiation (2007) 14, 963–972 & 2007 Nature Publishing Group All rights reserved 1350-9047/07 $30.00 www.nature.com/cdd Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells A-C Tosello-Trampont1,4, JM Kinchen1,2,4, E Brugnera1,3, LB Haney1, MO Hengartner2 and KS Ravichandran*,1 Removal of apoptotic cells is a dynamic process coordinated by ligands on apoptotic cells, and receptors and other signaling proteins on the phagocyte. One of the fundamental challenges is to understand how different phagocyte proteins form specific and functional complexes to orchestrate the recognition/removal of apoptotic cells. One evolutionarily conserved pathway involves the proteins cell death abnormal (CED)-2/chicken tumor virus no. 10 (CT10) regulator of kinase (Crk)II, CED-5/180 kDa protein downstream of chicken tumor virus no. 10 (Crk) (Dock180), CED-12/engulfment and migration (ELMO) and MIG-2/RhoG, leading to activation of the small GTPase CED-10/Rac and cytoskeletal remodeling to promote corpse uptake. Although the role of ELMO : Dock180 in regulating Rac activation has been well defined, the function of CED-2/CrkII in this complex is less well understood. Here, using functional studies in cell lines, we observe that a direct interaction between CrkII and Dock180 is not required for efficient removal of apoptotic cells. Similarly, mutants of CED-5 lacking the CED-2 interaction motifs could rescue engulfment and migration defects in CED-5 deficient worms. Mutants of CrkII and Dock180 that could not biochemically interact could colocalize in membrane ruffles. -

Disassembly of the Dying: Mechanisms and Functions
Review Disassembly of the Dying: Mechanisms and Functions 1 1, Georgia K. Atkin-Smith and Ivan K.H. Poon * The disassembly of an apoptotic cell into subcellular fragments, termed Trends apoptotic bodies (ApoBDs), is a hallmark of apoptosis. Although the genera- Apoptotic cell disassembly is a highly tion of ApoBDs is generally understood as being stochastic, it is becoming complex process regulated by a series of well-coordinated morphological increasingly clear that ApoBD formation is a highly regulated process involv- steps including apoptotic membrane ing distinct morphological steps and molecular factors. Functionally, ApoBDs blebbing, apoptotic protrusion forma- fi could facilitate the ef cient clearance of apoptotic material by surrounding tion, and fragmentation. phagocytes as well as mediate the transfer of biomolecules including micro- Plasma-membrane blebbing is not the RNAs and proteins between cells to aid in intercellular communications. sole process required for apoptotic Therefore, the formation of ApoBDs is an important process downstream body (ApoBD) formation, but mem- brane protrusions including microtu- from apoptotic cell death. We discuss here the mechanisms and functions bule spikes, apoptopodia, and of apoptotic cell disassembly. beaded apoptopodia may act in con- cert to aid the generation of ApoBDs. Cell Disassembly as a Key Downstream Process of Apoptotic Cell Death The mechanism of how apoptotic cells Billions of cells undergo apoptosis (a form of programmed cell death) daily as part of disassembly can determine the quantity and quality (size and contents) of physiological homeostasis [1]. At later stages of apoptosis, some cell types can generate ApoBDs. subcellular (1–5 mm) membrane-bound extracellular vesicles termed apoptotic bodies – (ApoBDs, see Glossary) [1 3]. -

Rho Guanine Nucleotide Exchange Factors: Regulators of Rho Gtpase Activity in Development and Disease
Oncogene (2014) 33, 4021–4035 & 2014 Macmillan Publishers Limited All rights reserved 0950-9232/14 www.nature.com/onc REVIEW Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease DR Cook1, KL Rossman2,3 and CJ Der1,2,3 The aberrant activity of Ras homologous (Rho) family small GTPases (20 human members) has been implicated in cancer and other human diseases. However, in contrast to the direct mutational activation of Ras found in cancer and developmental disorders, Rho GTPases are activated most commonly in disease by indirect mechanisms. One prevalent mechanism involves aberrant Rho activation via the deregulated expression and/or activity of Rho family guanine nucleotide exchange factors (RhoGEFs). RhoGEFs promote formation of the active GTP-bound state of Rho GTPases. The largest family of RhoGEFs is comprised of the Dbl family RhoGEFs with 70 human members. The multitude of RhoGEFs that activate a single Rho GTPase reflects the very specific role of each RhoGEF in controlling distinct signaling mechanisms involved in Rho activation. In this review, we summarize the role of Dbl RhoGEFs in development and disease, with a focus on Ect2 (epithelial cell transforming squence 2), Tiam1 (T-cell lymphoma invasion and metastasis 1), Vav and P-Rex1/2 (PtdIns(3,4,5)P3 (phosphatidylinositol (3,4,5)-triphosphate)-dependent Rac exchanger). Oncogene (2014) 33, 4021–4035; doi:10.1038/onc.2013.362; published online 16 September 2013 Keywords: Rac1; RhoA; Cdc42; guanine nucleotide exchange factors; cancer; -

Egfr Activates a Taz-Driven Oncogenic Program in Glioblastoma
EGFR ACTIVATES A TAZ-DRIVEN ONCOGENIC PROGRAM IN GLIOBLASTOMA by Minling Gao A thesis submitted to Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland March 2020 ©2020 Minling Gao All rights reserved Abstract Hyperactivated EGFR signaling is associated with about 45% of Glioblastoma (GBM), the most aggressive and lethal primary brain tumor in humans. However, the oncogenic transcriptional events driven by EGFR are still incompletely understood. We studied the role of the transcription factor TAZ to better understand master transcriptional regulators in mediating the EGFR signaling pathway in GBM. The transcriptional coactivator with PDZ- binding motif (TAZ) and its paralog gene, the Yes-associated protein (YAP) are two transcriptional co-activators that play important roles in multiple cancer types and are regulated in a context-dependent manner by various upstream signaling pathways, e.g. the Hippo, WNT and GPCR signaling. In GBM cells, TAZ functions as an oncogene that drives mesenchymal transition and radioresistance. This thesis intends to broaden our understanding of EGFR signaling and TAZ regulation in GBM. In patient-derived GBM cell models, EGF induced TAZ and its known gene targets through EGFR and downstream tyrosine kinases (ERK1/2 and STAT3). In GBM cells with EGFRvIII, an EGF-independent and constitutively active mutation, TAZ showed EGF- independent hyperactivation when compared to EGFRvIII-negative cells. These results revealed a novel EGFR-TAZ signaling axis in GBM cells. The second contribution of this thesis is that we performed next-generation sequencing to establish the first genome-wide map of EGF-induced TAZ target genes. -

A SPIKE1 Signaling Complex Controls Actin-Dependent Cell Morphogenesis Through the Heteromeric WAVE and ARP2/3 Complexes
A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes Dipanwita Basu*, Jie Le†, Taya Zakharova, Eileen L. Mallery, and Daniel B. Szymanski‡ Department of Agronomy, Purdue University, Lilly Hall, 915 W. State Street, West Lafayette, IN 47907-2054 Edited by Brian A. Larkins, University of Arizona, Tucson, AZ, and approved January 10, 2008 (received for review October 30, 2007) During morphogenesis, the actin cytoskeleton mediates cell-shape a conserved protein in both plants and animals and is a WAVE change in response to growth signals. In plants, actin filaments complex protein that directly interacts with active ROP/Rac (5, organize the cytoplasm in regions of polarized growth, and the 8). In cultured mammalian cells, Sra1 mediates the Rac- filamentous arrays can be highly dynamic. Small GTPase signaling dependent relocalization of the WAVE complex to leading edges proteins termed Rho of plants (ROP)/RAC control actin polymer- of lamellipodia (9), and in Arabidopsis, SRA1 and the WAVE ization. ROPs cycle between inactive GDP-bound and active GTP- complex positively regulate an actin filament-nucleating ma- bound forms, and it is the active form that interacts with effector chine, named the ARP2/3 complex (10). Plant and animal proteins to mediate cytoskeletal rearrangement and cell-shape WAVE complexes contain SCAR, a potent activator of ARP2/ change. A class of proteins termed guanine nucleotide exchange 3-dependent actin filament nucleation (8, 11). factors (GEFs) generate GTP–ROP and positively regulate ROP In both plant and animal systems, the proteins that generate signaling. However, in almost all experimental systems, it has the ROP/Rac signals and activate the WAVE–ARP2/3 pathway proven difficult to unravel the complex signaling pathways from are not known, and in general it has proven difficult to assign GEFs to the proteins that nucleate actin filaments. -

Regulation of Chemotaxis by the Orchestrated Activation of Ras, PI3K, and TOR Atsuo T
ARTICLE IN PRESS European Journal of Cell Biology 85 (2006) 873–895 www.elsevier.de/ejcb Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR Atsuo T. Sasakia,b, Richard A. Firtela,Ã aSection of Cell and Developmental Biology, Division of Biological Sciences, Center for Molecular Genetics, University of California, San Diego, Natural Sciences Building, Room 6316, 9500 Gilman Drive, La Jolla, CA 92093-0380, USA bDivision of Systems Biology, Harvard Medical School, Beth Israel Deaconess Medical Center, 77 Avenue Louis Pasteur NRB1052, Boston, MA 02115, USA Abstract Directed cell migration and cell polarity are crucial in many facets of biological processes. Cellular motility requires a complex array of signaling pathways, in which orchestrated cross-talk, a feedback loop, and multi-component signaling recur. Almost every signaling molecule requires several regulatory processes to be functionally activated, and a lack of a signaling molecule often leads to chemotaxis defects, suggesting an integral role for each component in the pathway. We outline our current understanding of the signaling event that regulates chemotaxis with an emphasis on recent findings associated with the Ras, PI3K, and target of rapamycin (TOR) pathways and the interplay of these pathways. Ras, PI3K, and TOR are known as key regulators of cellular growth. Deregulation of those pathways is associated with many human diseases, such as cancer, developmental disorders, and immunological deficiency. Recent studies in yeast, mammalian cells, and Dictyostelium discoideum reveal another critical role of Ras, PI3K, and TOR in regulating the actin cytoskeleton, cell polarity, and cellular movement. These findings shed light on the mechanism by which eukaryotic cells maintain cell polarity and directed cell movement, and also demonstrate that multiple steps in the signal transduction pathway coordinately regulate cell motility. -

Rho Gtpase Signalling Pathways in the Morphological Changes Associated with Apoptosis
Cell Death and Differentiation (2002) 9, 493 ± 504 ã 2002 Nature Publishing Group All rights reserved 1350-9047/02 $25.00 www.nature.com/cdd Review Rho GTPase signalling pathways in the morphological changes associated with apoptosis ML Coleman1 and MF Olson*,1 membrane blebs and apoptotic bodies (reviewed in refer- ence3). Ultimately, the dead cell is packaged into membrane- 1 Cancer Research Campaign Centre for Cell and Molecular Biology, Institute of clad apoptotic bodies that facilitate uptake by neighbouring Cancer Research, 237 Fulham Road, London SW3 6JB, UK cells or by specialised phagocytic cells. Dynamic re- * Corresponding author: MF Olson, Abramson Family Cancer Research arrangements of the actin cytoskeleton in the phagocyte are Institute, Room 411, BRB II/III, 421 Curie Boulevard, University of required for the internalisation of apoptotic cell fragments. Pennsylvania, Philadelphia, PA 19104-6160, USA. Tel: +1 215 746 6798; Fax: +1 215 746 5525; E-mail: [email protected] Recent research has revealed the importance of signal transduction pathways controlled by Rho family GTPases in Received 23.8.01; revised 26.10.01; accepted 5.11.01 regulating the marked changes in cell morphology observed Edited by M Piacentini in the processes of apoptosis and phagocytosis. The Rho GTPases are a family of proteins (RhoA, RhoB, RhoC, RhoD, RhoE/Rnd3, RhoG, RhoH/TTF, Rnd1, Rnd2, Abstract Rac1, Rac2, Rac3, Cdc42/G25K, Wrch-1, TC10, TCL, Chp, Rif) that act as molecular switches in intracellular signal The killing and removal of superfluous cells is an important 4 step during embryonic development, tissue homeostasis, transduction pathways (reviewed in reference ). -

Temporal Deregulation of Genes and Micrornas in Neurons During Prion-Induced Neurodegeneration
Temporal Deregulation of Genes and MicroRNAs in Neurons during Prion-Induced Neurodegeneration By ANNA MAJER A Thesis Submitted to the Faculty of Graduate Studies In Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY Department of Medical Microbiology College of Medicine, Faculty of Health Sciences University of Manitoba Winnipeg, Manitoba © Anna Majer, October 2015 TABLE OF CONTENTS ABSTRACT .................................................................................................................................... 8 ACKNOWLEDGMENTS ............................................................................................................ 10 ABBREVIATIONS ...................................................................................................................... 12 LIST OF FIGURES ...................................................................................................................... 15 LIST OF TABLES ........................................................................................................................ 19 1.0 INTRODUCTION .................................................................................................................. 20 1.1 Common Elements of Neurodegenerative Diseases ........................................................... 21 1.2 Rodent Models for Neurodegeneration ............................................................................... 22 1.3 Prion Diseases .................................................................................................................... -

The RHO Family Gtpases: Mechanisms of Regulation and Signaling
cells Review The RHO Family GTPases: Mechanisms of Regulation and Signaling Niloufar Mosaddeghzadeh and Mohammad Reza Ahmadian * Institute of Biochemistry and Molecular Biology II, Medical Faculty of the Heinrich Heine University, Universitätsstrasse 1, Building 22.03.05, 40225 Düsseldorf, Germany; [email protected] * Correspondence: [email protected] Abstract: Much progress has been made toward deciphering RHO GTPase functions, and many studies have convincingly demonstrated that altered signal transduction through RHO GTPases is a recurring theme in the progression of human malignancies. It seems that 20 canonical RHO GTPases are likely regulated by three GDIs, 85 GEFs, and 66 GAPs, and eventually interact with >70 downstream effectors. A recurring theme is the challenge in understanding the molecular determinants of the specificity of these four classes of interacting proteins that, irrespective of their functions, bind to common sites on the surface of RHO GTPases. Identified and structurally verified hotspots as functional determinants specific to RHO GTPase regulation by GDIs, GEFs, and GAPs as well as signaling through effectors are presented, and challenges and future perspectives are discussed. Keywords: CDC42; effectors; RAC1; RHOA; RHOGAP; RHOGDI; RHOGEF; RHO signaling 1. Introduction Citation: Mosaddeghzadeh, N.; The RHO (RAS homolog) family is an integral part of the RAS superfamily of guanine Ahmadian, M.R. The RHO Family nucleotide-binding proteins. RHO family proteins are crucial for several reasons: (i) ap- GTPases: Mechanisms of Regulation proximately 1% of the human genome encodes proteins that either regulate or are regulated and Signaling. Cells 2021, 10, 1831. by direct interaction with RHO proteins; (ii) they control almost all fundamental cellular https://doi.org/10.3390/cells10071831 processes in eukaryotes including morphogenesis, polarity, movement, cell division, gene expression, and cytoskeleton reorganization [1]; and (iii) they are associated with a series Academic Editor: Bor Luen Tang of human diseases (Figure1)[2].