Rho Guanine Nucleotide Exchange Factors: Regulators of Rho Gtpase Activity in Development and Disease
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Cytoplasmic Expression of Epithelial Cell Transforming Sequence 2 in Lung Adenocarcinoma and Its Implications for Malignant Progression
Laboratory Investigation (2019) 99:551–567 https://doi.org/10.1038/s41374-018-0142-4 ARTICLE Cytoplasmic expression of epithelial cell transforming sequence 2 in lung adenocarcinoma and its implications for malignant progression 1 2 3 1 2 2 Zeinab Kosibaty ● Yoshihiko Murata ● Yuko Minami ● Tomoko Dai ● Junko Kano ● Ryota Matsuoka ● 2 2 Noriyuki Nakano ● Masayuki Noguchi Received: 8 March 2018 / Revised: 14 August 2018 / Accepted: 20 August 2018 / Published online: 12 December 2018 © United States & Canadian Academy of Pathology 2018 Abstract Epithelial cell transforming sequence 2 (ECT2), a guanine nucleotide exchange factor, is predominantly localized in the nucleus of non-transformed cells and functions to regulate cytokinesis. ECT2 is also localized in the cytoplasm of cancer cells. Aberrant cytoplasmic expression of ECT2 is thought to drive tumor growth and invasion. In this study, we investigated the cytoplasmic expression of ECT2 and its prognostic and biological significance in lung adenocarcinoma. Western blotting of cellular fractions from the nucleus and cytoplasm was performed to determine the subcellular localization of ECT2 in lung adenocarcinoma cell lines. The cytoplasmic expression of ECT2 in 167 lung fi 1234567890();,: 1234567890();,: adenocarcinomas was evaluated by immunohistochemistry and its clinical signi cance was examined using Kaplan–Meier curves and Cox regression analysis. Scraping cytology specimens of 13 fresh lung adenocarcinomas were used to assess the subcellular localization of ECT2 and its phosphorylation at Thr790 (P-ECT2(T790)). We found that ECT2 was localized in both the nucleus and cytoplasm of lung adenocarcinoma cell lines and tumor tissues. Cytoplasmic expression of ECT2 was detected by immunohistochemistry in 83 (50%) of the lung adenocarcinomas, and was found to increase during cancer progression. -

Genome-Wide Analysis of DNA Methylation Differences in Muscle and Fat from Monozygotic Twins Discordant for Type 2 Diabetes
Genome-Wide Analysis of DNA Methylation Differences in Muscle and Fat from Monozygotic Twins Discordant for Type 2 Diabetes Rasmus Ribel-Madsen1,2*, Mario F. Fraga3, Stine Jacobsen1, Jette Bork-Jensen1, Ester Lara4, Vincenzo Calvanese4, Agustin F. Fernandez5, Martin Friedrichsen1, Birgitte F. Vind6, Kurt Højlund6, Henning Beck-Nielsen6, Manel Esteller7, Allan Vaag1,8, Pernille Poulsen9 1 Steno Diabetes Center, Gentofte, Denmark, 2 Section of Metabolic Genetics, The Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark, 3 Department of Immunology and Oncology, National Centre for Biotechnology, CNB-CSIC, Madrid, Spain, 4 Department of Cancer Epigenetics, Spanish National Cancer Research Centre, Madrid, Spain, 5 Cancer Epigenetics Laboratory, Instituto Universitario de Oncologia del Principado de Asturias, HUCA, Universidad de Oviedo, Oviedo, Spain, 6 Department of Endocrinology, Odense University Hospital, Odense, Denmark, 7 Cancer Epigenetics and Biology Program (PEBC), Bellvitge Biomedical Research Institute (IDIBELL), L’Hospitalet de Llobregat, Barcelona, Spain, 8 Department of Endocrinology, Rigshospitalet, Copenhagen, Denmark, 9 Novo Nordisk A/S, Søborg, Denmark Abstract Background: Monozygotic twins discordant for type 2 diabetes constitute an ideal model to study environmental contributions to type 2 diabetic traits. We aimed to examine whether global DNA methylation differences exist in major glucose metabolic tissues from these twins. Methodology/Principal Findings: Skeletal muscle (n = 11 pairs) and subcutaneous adipose tissue (n = 5 pairs) biopsies were collected from 53–80 year-old monozygotic twin pairs discordant for type 2 diabetes. DNA methylation was measured by microarrays at 26,850 cytosine-guanine dinucleotide (CpG) sites in the promoters of 14,279 genes. -
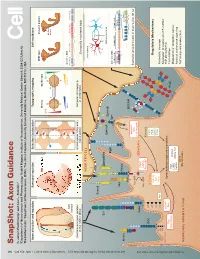
Snapshot: Axon Guidance Pasterkamp R
494 1 Cell Cell ??? SnapShot: Axon Guidance 153 SnapShot: XXXXXXXXXXXXXXXXXXXXXXXXXX 1 2 , ??MONTH?? ??DATE??, 200? ©200? Elsevier Inc. 200?©200? ElsevierInc. , ??MONTH?? ??DATE??, DOI R. Jeroen Pasterkamp and Alex L. Kolodkin , April11, 2013©2013Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.03.031 AUTHOR XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 1 AFFILIATIONDepartment of XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX Neuroscience and Pharmacology, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, 3584 CG Utrecht, The Netherlands; 2Department of Neuroscience, HHMI, The Johns Hopkins University School of Medicine, Baltimore, MD 21212, USA Axon attraction and repulsion Surround repulsion Selective fasciculation Topographic mapping Self-avoidance Wild-type Dscam1 mutant Sema3A A Retina SC/Tectum EphB EphrinB P D D Mutant T A neuron N P A V V Genomic DNA P EphA EphrinA Slit Exon 4 (12) Exon 6 (48) Exon 9 (33) Exon 17 (2) Netrin Commissural axon guidance Surround repulsion of peripheral Grasshopper CNS axon Retinotectal mapping at the CNS midline nerves in vertebrates fasciculation in vertebrates Drosophila mushroom body XXXXXXXXX NEURITE/CELL Isoneuronal Sema3 Slit Sema1/4-6 Heteroneuronal EphrinA EphrinB FasII Eph Genomic DNA Pcdh-α (14) Pcdh-β (22) Pcdh-γ (22) Variable Con Variable Con Nrp Plexin ** *** Netrin LAMELLIPODIA Con DSCAM ephexin Starburst amacrine cells in mammalian retina Ras-GTP Vav See online version for legend and references. α-chimaerin GEFs/GAPs Robo FARP Ras-GDP LARG RhoGEF Kinases DCC cc0 GTPases PKA cc1 FAK Regulatory Mechanisms See online versionfor??????. Cdc42 GSK3 cc2 Rac PI3K P1 Rho P2 cc3 Abl Proteolytic cleavage P3 Regulation of expression (TF, miRNA, FILOPODIA srGAP Cytoskeleton regulatory proteins multiple isoforms) Sos Trio Pcdh Cis inhibition DOCK180 PAK ROCK Modulation of receptors’ output LIMK Myosin-II Colin Forward and reverse signaling Actin Trafcking and endocytosis NEURONAL GROWTH CONE Microtubules SnapShot: Axon Guidance R. -

AN INVESTIGATION of the ROLE of PAK6 in TUMORIGENESIS By
AN INVESTIGATION OF THE ROLE OF PAK6 IN TUMORIGENESIS by JoAnn Roberts A Thesis Submitted to the Faculty of The Charles E. Schmidt College of Medicine In Partial Fulfillment of the Requirements for the Degree of Master of Science Florida Atlantic University Boca Raton, Florida August 2012 ACKNOWLEDGMENTS This material is based upon work supported by the National Science Foundation under Grant No. DGE: 0638662. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. I would like to thank and acknowledge my thesis advisor, Dr. Michael Lu, for his support and guidance throughout the writing of this thesis and design of experiments in this manuscript. I would also like to thank my colleagues for assistance in various trouble-shooting circumstances. Last, but certainly not least, I would like to thank my family and friends for their support in the pursuit of my graduate studies. iii ABSTRACT Author: JoAnn Roberts Title: An Investigation of the Role of PAK6 in Tumorigenesis Institution: Florida Atlantic University Thesis Advisor: Dr. Michael Lu Degree: Master of Science Year: 2012 The function and role of PAK6, a serine/threonine kinase, in cancer progression has not yet been clearly identified. Several studies reveal that PAK6 may participate in key changes contributing to cancer progression such as cell survival, cell motility, and invasiveness. Based on the membrane localization of PAK6 in prostate and breast cancer cells, we speculated that PAK6 plays a role in cancer progression cells by localizing on the membrane and modifying proteins linked to motility and proliferation. -

Regulation of Cdc42 and Its Effectors in Epithelial Morphogenesis Franck Pichaud1,2,*, Rhian F
© 2019. Published by The Company of Biologists Ltd | Journal of Cell Science (2019) 132, jcs217869. doi:10.1242/jcs.217869 REVIEW SUBJECT COLLECTION: ADHESION Regulation of Cdc42 and its effectors in epithelial morphogenesis Franck Pichaud1,2,*, Rhian F. Walther1 and Francisca Nunes de Almeida1 ABSTRACT An overview of Cdc42 Cdc42 – a member of the small Rho GTPase family – regulates cell Cdc42 was discovered in yeast and belongs to a large family of small – polarity across organisms from yeast to humans. It is an essential (20 30 kDa) GTP-binding proteins (Adams et al., 1990; Johnson regulator of polarized morphogenesis in epithelial cells, through and Pringle, 1990). It is part of the Ras-homologous Rho subfamily coordination of apical membrane morphogenesis, lumen formation and of GTPases, of which there are 20 members in humans, including junction maturation. In parallel, work in yeast and Caenorhabditis elegans the RhoA and Rac GTPases, (Hall, 2012). Rho, Rac and Cdc42 has provided important clues as to how this molecular switch can homologues are found in all eukaryotes, except for plants, which do generate and regulate polarity through localized activation or inhibition, not have a clear homologue for Cdc42. Together, the function of and cytoskeleton regulation. Recent studies have revealed how Rho GTPases influences most, if not all, cellular processes. important and complex these regulations can be during epithelial In the early 1990s, seminal work from Alan Hall and his morphogenesis. This complexity is mirrored by the fact that Cdc42 can collaborators identified Rho, Rac and Cdc42 as main regulators of exert its function through many effector proteins. -

Cell Division Symmetry Control and Cancer Stem Cells
AIMS Molecular Science, 7(2): 82–98. DOI: 10.3934/molsci.2020006 Received: 15 February 2020 Accepted: 26 April 2020 Published: 06 May 2020 http://www.aimspress.com/journal/Molecular Review Cell division symmetry control and cancer stem cells Sreemita Majumdar and Song-Tao Liu* Department of Biological Sciences, University of Toledo, Toledo, OH 43606, USA * Correspondence: Email: [email protected]; Tel: +14195307853. Table S1. Genes encoding polarity and fate-determinant proteins involved in asymmetric cell division. C. elegans1 D. melanogaster 1 Mammals1 Description2 Associated with/ Interactors 3 Cellular Localization (mammalian cell)4 Serine/threonine protein microtubule-associated protein cell membrane, peripheral and lateral, par-1 par-1 MARK1/2/3/4 kinase MAPT/TAU cytoplasm, dendrite RING, Lipid binding par-2 - - domain PDZ for membrane, cell junction, adherens junction, cell cortex, par-3 baz PARD3 Oligomerization domain at actin, PARD6 endomembrane system, NTD Continued on next page 2 C. elegans1 D. melanogaster 1 Mammals1 Description2 Associated with/ Interactors 3 Cellular Localization (mammalian cell)4 Serine/threonine-protein nucleus, mitochondria, cytoplasm, par-4 Lkb1 STK11/LKB1 STRAD complex kinase membrane 14-3-3 domain binding par-5 14-3-3 YWHAB phosphoserine/ adapter to many proteins cytoplasm phosphothreonine motif cell membrane, centriolar satellite, actin par-6 par-6 PARD6A/B/G PB1, CRIB, PDZ PARD3 cytoskeleton,centrosome, cytoplasm ,ruffles PARD3, and a PARD6 protein PB1, AGC-Kinase (PARD6A, PARD6B or PARD6G) pkc-3 aPKC PRKCI/Z domain, DAG binding, cytoplasm, nucleus, membrane and a GTPase protein (CDC42 or Zinc finger domain RAC1), LLGL1,ECT2 LRR and PDZ protein Cadherin, Scrib-APC-beta-catenin nucleoplasm, basolateral plasma membrane, let-413 scrib SCRIB family. -

Regulation of the Mammalian Target of Rapamycin Complex 2 (Mtorc2)
Regulation of the Mammalian Target Of Rapamycin Complex 2 (mTORC2) Inauguraldissertation Zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Klaus-Dieter Molle aus Heilbronn, Deutschland Basel, 2006 Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät Auf Antrag von Prof. Michael N. Hall und Prof. Markus Affolter. Basel, den 21.11.2006 Prof. Hans-Peter Hauri Dekan Summary The growth controlling mammalian Target of Rapamycin (mTOR) is a conserved Ser/Thr kinase found in two structurally and functionally distinct complexes, mTORC1 and mTORC2. The tumor suppressor TSC1-TSC2 complex inhibits mTORC1 by acting on the small GTPase Rheb, but the role of TSC1-TSC2 and Rheb in the regulation of mTORC2 is unclear. Here we examined the role of TSC1-TSC2 in the regulation of mTORC2 in human embryonic kidney 293 cells. Induced knockdown of TSC1 and TSC2 (TSC1/2) stimulated mTORC2-dependent actin cytoskeleton organization and Paxillin phosphorylation. Furthermore, TSC1/2 siRNA increased mTORC2-dependent Ser473 phosphorylation of plasma membrane bound, myristoylated Akt/PKB. This suggests that loss of Akt/PKB Ser473 phosphorylation in TSC mutant cells, as reported previously, is due to inhibition of Akt/PKB localization rather than inhibition of mTORC2 activity. Amino acids and overexpression of Rheb failed to stimulate mTORC2 signaling. Thus, TSC1-TSC2 also inhibits mTORC2, but possibly independently of Rheb. Our results suggest that mTORC2 hyperactivation may contribute to the pathophysiology of diseases such as cancer and Tuberous Sclerosis Complex. i Acknowledgement During my PhD studies in the Biozentrum I received a lot of support from many people around me who I mention here to express my gratefulness. -

The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination
The Journal of Neuroscience, August 31, 2011 • 31(35):12579–12592 • 12579 Cellular/Molecular The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination Junji Yamauchi,1,3,5 Yuki Miyamoto,1 Hajime Hamasaki,1,3 Atsushi Sanbe,1 Shinji Kusakawa,1 Akane Nakamura,2 Hideki Tsumura,2 Masahiro Maeda,4 Noriko Nemoto,6 Katsumasa Kawahara,5 Tomohiro Torii,1 and Akito Tanoue1 1Department of Pharmacology and 2Laboratory Animal Resource Facility, National Research Institute for Child Health and Development, Setagaya, Tokyo 157-8535, Japan, 3Department of Biological Sciences, Tokyo Institute of Technology, Midori, Yokohama 226-8501, Japan, 4IBL, Ltd., Fujioka, Gumma 375-0005, Japan, and 5Department of Physiology and 6Bioimaging Research Center, Kitasato University School of Medicine, Sagamihara, Kanagawa 252-0374, Japan In development of the peripheral nervous system, Schwann cells proliferate, migrate, and ultimately differentiate to form myelin sheath. In all of the myelination stages, Schwann cells continuously undergo morphological changes; however, little is known about their underlying molecular mechanisms. We previously cloned the dock7 gene encoding the atypical Rho family guanine-nucleotide exchange factor (GEF) and reported the positive role of Dock7, the target Rho GTPases Rac/Cdc42, and the downstream c-Jun N-terminal kinase in Schwann cell migration (Yamauchi et al., 2008). We investigated the role of Dock7 in Schwann cell differentiation and myelination. Knockdown of Dock7 by the specific small interfering (si)RNA in primary Schwann cells promotes dibutyryl cAMP-induced morpholog- ical differentiation, indicating the negative role of Dock7 in Schwann cell differentiation. It also results in a shorter duration of activation of Rac/Cdc42 and JNK, which is the negative regulator of myelination, and the earlier activation of Rho and Rho-kinase, which is the positive regulator of myelination. -

Related Malignant Phenotypes in the Nf1-Deficient MPNST
Published OnlineFirst February 19, 2013; DOI: 10.1158/1541-7786.MCR-12-0593 Molecular Cancer Genomics Research RAS/MEK–Independent Gene Expression Reveals BMP2- Related Malignant Phenotypes in the Nf1-Deficient MPNST Daochun Sun1, Ramsi Haddad2,3, Janice M. Kraniak2, Steven D. Horne1, and Michael A. Tainsky1,2 Abstract Malignant peripheral nerve sheath tumor (MPNST) is a type of soft tissue sarcoma that occurs in carriers of germline mutations in Nf1 gene as well as sporadically. Neurofibromin, encoded by the Nf1 gene, functions as a GTPase-activating protein (GAP) whose mutation leads to activation of wt-RAS and mitogen-activated protein kinase (MAPK) signaling in neurofibromatosis type I (NF1) patients' tumors. However, therapeutic targeting of RAS and MAPK have had limited success in this disease. In this study, we modulated NRAS, mitogen-activated protein/extracellular signal–regulated kinase (MEK)1/2, and neurofibromin levels in MPNST cells and determined gene expression changes to evaluate the regulation of signaling pathways in MPNST cells. Gene expression changes due to neurofibromin modulation but independent of NRAS and MEK1/2 regulation in MPNST cells indicated bone morphogenetic protein 2 (Bmp2) signaling as a key pathway. The BMP2-SMAD1/5/8 pathway was activated in NF1-associated MPNST cells and inhibition of BMP2 signaling by LDN-193189 or short hairpin RNA (shRNA) to BMP2 decreased the motility and invasion of NF1-associated MPNST cells. The pathway-specific gene changes provide a greater understanding of the complex role of neurofibromin in MPNST pathology and novel targets for drug discovery. Mol Cancer Res; 11(6); 616–27. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

A Rac/Cdc42 Exchange Factor Complex Promotes Formation of Lateral filopodia and Blood Vessel Lumen Morphogenesis
ARTICLE Received 1 Oct 2014 | Accepted 26 Apr 2015 | Published 1 Jul 2015 DOI: 10.1038/ncomms8286 OPEN A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis Sabu Abraham1,w,*, Margherita Scarcia2,w,*, Richard D. Bagshaw3,w,*, Kathryn McMahon2,w, Gary Grant2, Tracey Harvey2,w, Maggie Yeo1, Filomena O.G. Esteves2, Helene H. Thygesen2,w, Pamela F. Jones4, Valerie Speirs2, Andrew M. Hanby2, Peter J. Selby2, Mihaela Lorger2, T. Neil Dear4,w, Tony Pawson3,z, Christopher J. Marshall1 & Georgia Mavria2 During angiogenesis, Rho-GTPases influence endothelial cell migration and cell–cell adhesion; however it is not known whether they control formation of vessel lumens, which are essential for blood flow. Here, using an organotypic system that recapitulates distinct stages of VEGF-dependent angiogenesis, we show that lumen formation requires early cytoskeletal remodelling and lateral cell–cell contacts, mediated through the RAC1 guanine nucleotide exchange factor (GEF) DOCK4 (dedicator of cytokinesis 4). DOCK4 signalling is necessary for lateral filopodial protrusions and tubule remodelling prior to lumen formation, whereas proximal, tip filopodia persist in the absence of DOCK4. VEGF-dependent Rac activation via DOCK4 is necessary for CDC42 activation to signal filopodia formation and depends on the activation of RHOG through the RHOG GEF, SGEF. VEGF promotes interaction of DOCK4 with the CDC42 GEF DOCK9. These studies identify a novel Rho-family GTPase activation cascade for the formation of endothelial cell filopodial protrusions necessary for tubule remodelling, thereby influencing subsequent stages of lumen morphogenesis. 1 Institute of Cancer Research, Division of Cancer Biology, 237 Fulham Road, London SW3 6JB, UK.