A SPIKE1 Signaling Complex Controls Actin-Dependent Cell Morphogenesis Through the Heteromeric WAVE and ARP2/3 Complexes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
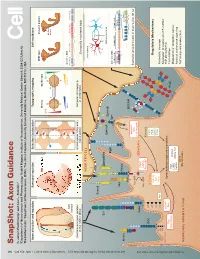
Snapshot: Axon Guidance Pasterkamp R
494 1 Cell Cell ??? SnapShot: Axon Guidance 153 SnapShot: XXXXXXXXXXXXXXXXXXXXXXXXXX 1 2 , ??MONTH?? ??DATE??, 200? ©200? Elsevier Inc. 200?©200? ElsevierInc. , ??MONTH?? ??DATE??, DOI R. Jeroen Pasterkamp and Alex L. Kolodkin , April11, 2013©2013Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.03.031 AUTHOR XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 1 AFFILIATIONDepartment of XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX Neuroscience and Pharmacology, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, 3584 CG Utrecht, The Netherlands; 2Department of Neuroscience, HHMI, The Johns Hopkins University School of Medicine, Baltimore, MD 21212, USA Axon attraction and repulsion Surround repulsion Selective fasciculation Topographic mapping Self-avoidance Wild-type Dscam1 mutant Sema3A A Retina SC/Tectum EphB EphrinB P D D Mutant T A neuron N P A V V Genomic DNA P EphA EphrinA Slit Exon 4 (12) Exon 6 (48) Exon 9 (33) Exon 17 (2) Netrin Commissural axon guidance Surround repulsion of peripheral Grasshopper CNS axon Retinotectal mapping at the CNS midline nerves in vertebrates fasciculation in vertebrates Drosophila mushroom body XXXXXXXXX NEURITE/CELL Isoneuronal Sema3 Slit Sema1/4-6 Heteroneuronal EphrinA EphrinB FasII Eph Genomic DNA Pcdh-α (14) Pcdh-β (22) Pcdh-γ (22) Variable Con Variable Con Nrp Plexin ** *** Netrin LAMELLIPODIA Con DSCAM ephexin Starburst amacrine cells in mammalian retina Ras-GTP Vav See online version for legend and references. α-chimaerin GEFs/GAPs Robo FARP Ras-GDP LARG RhoGEF Kinases DCC cc0 GTPases PKA cc1 FAK Regulatory Mechanisms See online versionfor??????. Cdc42 GSK3 cc2 Rac PI3K P1 Rho P2 cc3 Abl Proteolytic cleavage P3 Regulation of expression (TF, miRNA, FILOPODIA srGAP Cytoskeleton regulatory proteins multiple isoforms) Sos Trio Pcdh Cis inhibition DOCK180 PAK ROCK Modulation of receptors’ output LIMK Myosin-II Colin Forward and reverse signaling Actin Trafcking and endocytosis NEURONAL GROWTH CONE Microtubules SnapShot: Axon Guidance R. -

Regulation of the Mammalian Target of Rapamycin Complex 2 (Mtorc2)
Regulation of the Mammalian Target Of Rapamycin Complex 2 (mTORC2) Inauguraldissertation Zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Klaus-Dieter Molle aus Heilbronn, Deutschland Basel, 2006 Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät Auf Antrag von Prof. Michael N. Hall und Prof. Markus Affolter. Basel, den 21.11.2006 Prof. Hans-Peter Hauri Dekan Summary The growth controlling mammalian Target of Rapamycin (mTOR) is a conserved Ser/Thr kinase found in two structurally and functionally distinct complexes, mTORC1 and mTORC2. The tumor suppressor TSC1-TSC2 complex inhibits mTORC1 by acting on the small GTPase Rheb, but the role of TSC1-TSC2 and Rheb in the regulation of mTORC2 is unclear. Here we examined the role of TSC1-TSC2 in the regulation of mTORC2 in human embryonic kidney 293 cells. Induced knockdown of TSC1 and TSC2 (TSC1/2) stimulated mTORC2-dependent actin cytoskeleton organization and Paxillin phosphorylation. Furthermore, TSC1/2 siRNA increased mTORC2-dependent Ser473 phosphorylation of plasma membrane bound, myristoylated Akt/PKB. This suggests that loss of Akt/PKB Ser473 phosphorylation in TSC mutant cells, as reported previously, is due to inhibition of Akt/PKB localization rather than inhibition of mTORC2 activity. Amino acids and overexpression of Rheb failed to stimulate mTORC2 signaling. Thus, TSC1-TSC2 also inhibits mTORC2, but possibly independently of Rheb. Our results suggest that mTORC2 hyperactivation may contribute to the pathophysiology of diseases such as cancer and Tuberous Sclerosis Complex. i Acknowledgement During my PhD studies in the Biozentrum I received a lot of support from many people around me who I mention here to express my gratefulness. -

The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination
The Journal of Neuroscience, August 31, 2011 • 31(35):12579–12592 • 12579 Cellular/Molecular The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination Junji Yamauchi,1,3,5 Yuki Miyamoto,1 Hajime Hamasaki,1,3 Atsushi Sanbe,1 Shinji Kusakawa,1 Akane Nakamura,2 Hideki Tsumura,2 Masahiro Maeda,4 Noriko Nemoto,6 Katsumasa Kawahara,5 Tomohiro Torii,1 and Akito Tanoue1 1Department of Pharmacology and 2Laboratory Animal Resource Facility, National Research Institute for Child Health and Development, Setagaya, Tokyo 157-8535, Japan, 3Department of Biological Sciences, Tokyo Institute of Technology, Midori, Yokohama 226-8501, Japan, 4IBL, Ltd., Fujioka, Gumma 375-0005, Japan, and 5Department of Physiology and 6Bioimaging Research Center, Kitasato University School of Medicine, Sagamihara, Kanagawa 252-0374, Japan In development of the peripheral nervous system, Schwann cells proliferate, migrate, and ultimately differentiate to form myelin sheath. In all of the myelination stages, Schwann cells continuously undergo morphological changes; however, little is known about their underlying molecular mechanisms. We previously cloned the dock7 gene encoding the atypical Rho family guanine-nucleotide exchange factor (GEF) and reported the positive role of Dock7, the target Rho GTPases Rac/Cdc42, and the downstream c-Jun N-terminal kinase in Schwann cell migration (Yamauchi et al., 2008). We investigated the role of Dock7 in Schwann cell differentiation and myelination. Knockdown of Dock7 by the specific small interfering (si)RNA in primary Schwann cells promotes dibutyryl cAMP-induced morpholog- ical differentiation, indicating the negative role of Dock7 in Schwann cell differentiation. It also results in a shorter duration of activation of Rac/Cdc42 and JNK, which is the negative regulator of myelination, and the earlier activation of Rho and Rho-kinase, which is the positive regulator of myelination. -

A Rac/Cdc42 Exchange Factor Complex Promotes Formation of Lateral filopodia and Blood Vessel Lumen Morphogenesis
ARTICLE Received 1 Oct 2014 | Accepted 26 Apr 2015 | Published 1 Jul 2015 DOI: 10.1038/ncomms8286 OPEN A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis Sabu Abraham1,w,*, Margherita Scarcia2,w,*, Richard D. Bagshaw3,w,*, Kathryn McMahon2,w, Gary Grant2, Tracey Harvey2,w, Maggie Yeo1, Filomena O.G. Esteves2, Helene H. Thygesen2,w, Pamela F. Jones4, Valerie Speirs2, Andrew M. Hanby2, Peter J. Selby2, Mihaela Lorger2, T. Neil Dear4,w, Tony Pawson3,z, Christopher J. Marshall1 & Georgia Mavria2 During angiogenesis, Rho-GTPases influence endothelial cell migration and cell–cell adhesion; however it is not known whether they control formation of vessel lumens, which are essential for blood flow. Here, using an organotypic system that recapitulates distinct stages of VEGF-dependent angiogenesis, we show that lumen formation requires early cytoskeletal remodelling and lateral cell–cell contacts, mediated through the RAC1 guanine nucleotide exchange factor (GEF) DOCK4 (dedicator of cytokinesis 4). DOCK4 signalling is necessary for lateral filopodial protrusions and tubule remodelling prior to lumen formation, whereas proximal, tip filopodia persist in the absence of DOCK4. VEGF-dependent Rac activation via DOCK4 is necessary for CDC42 activation to signal filopodia formation and depends on the activation of RHOG through the RHOG GEF, SGEF. VEGF promotes interaction of DOCK4 with the CDC42 GEF DOCK9. These studies identify a novel Rho-family GTPase activation cascade for the formation of endothelial cell filopodial protrusions necessary for tubule remodelling, thereby influencing subsequent stages of lumen morphogenesis. 1 Institute of Cancer Research, Division of Cancer Biology, 237 Fulham Road, London SW3 6JB, UK. -

Targeting PH Domain Proteins for Cancer Therapy
The Texas Medical Center Library DigitalCommons@TMC The University of Texas MD Anderson Cancer Center UTHealth Graduate School of The University of Texas MD Anderson Cancer Biomedical Sciences Dissertations and Theses Center UTHealth Graduate School of (Open Access) Biomedical Sciences 12-2018 Targeting PH domain proteins for cancer therapy Zhi Tan Follow this and additional works at: https://digitalcommons.library.tmc.edu/utgsbs_dissertations Part of the Bioinformatics Commons, Medicinal Chemistry and Pharmaceutics Commons, Neoplasms Commons, and the Pharmacology Commons Recommended Citation Tan, Zhi, "Targeting PH domain proteins for cancer therapy" (2018). The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access). 910. https://digitalcommons.library.tmc.edu/utgsbs_dissertations/910 This Dissertation (PhD) is brought to you for free and open access by the The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences at DigitalCommons@TMC. It has been accepted for inclusion in The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access) by an authorized administrator of DigitalCommons@TMC. For more information, please contact [email protected]. TARGETING PH DOMAIN PROTEINS FOR CANCER THERAPY by Zhi Tan Approval page APPROVED: _____________________________________________ Advisory Professor, Shuxing Zhang, Ph.D. _____________________________________________ -

Crit. Rev. in Biochem. and Molec. Biol. 2011
Critical Reviews in Biochemistry and Molecular Biology Critical Reviews in Biochemistry and Molecular Biology, 2011, 1–21, Early Online 2011 © 2011 Informa Healthcare USA, Inc. ISSN 1040-9238 print/ISSN 1549-7798 online 00 DOI: 10.3109/10409238.2011.618113 00 000 REVIEW 000 18 June 2011 Mammalian TOR signaling to the AGC kinases 15 August 2011 Bing Su1 and Estela Jacinto2 24 August 2011 1Department of Immunobiology and The Vascular Biology and Therapeutics Program, Yale University School of Medicine, New Haven, CT, USA and 2Department of Physiology and Biophysics, UMDNJ-RWJMS, Piscataway, NJ, USA 1040-9238 1549-7798 Abstract The mechanistic (or mammalian) target of rapamycin (mTOR), an evolutionarily conserved protein kinase, orchestrates © 2011 Informa Healthcare USA, Inc. cellular responses to growth, metabolic and stress signals. mTOR processes various extracellular and intracellular inputs as part of two mTOR protein complexes, mTORC1 or mTORC2. The mTORCs have numerous cellular targets but 10.3109/10409238.2011.618113 members of a family of protein kinases, the protein kinase (PK)A/PKG/PKC (AGC) family are the best characterized direct mTOR substrates. The AGC kinases control multiple cellular functions and deregulation of many members of BBMG this family underlies numerous pathological conditions. mTOR phosphorylates conserved motifs in these kinases to allosterically augment their activity, influence substrate specificity, and promote protein maturation and 618113 stability. Activation of AGC kinases in turn triggers the phosphorylation of diverse, often overlapping, targets that ultimately control cellular response to a wide spectrum of stimuli. This review will highlight recent findings on how mTOR regulates AGC kinases and how mTOR activity is feedback regulated by these kinases. -

Cldn19 Clic2 Clmp Cln3
NewbornDx™ Advanced Sequencing Evaluation When time to diagnosis matters, the NewbornDx™ Advanced Sequencing Evaluation from Athena Diagnostics delivers rapid, 5- to 7-day results on a targeted 1,722-genes. A2ML1 ALAD ATM CAV1 CLDN19 CTNS DOCK7 ETFB FOXC2 GLUL HOXC13 JAK3 AAAS ALAS2 ATP1A2 CBL CLIC2 CTRC DOCK8 ETFDH FOXE1 GLYCTK HOXD13 JUP AARS2 ALDH18A1 ATP1A3 CBS CLMP CTSA DOK7 ETHE1 FOXE3 GM2A HPD KANK1 AASS ALDH1A2 ATP2B3 CC2D2A CLN3 CTSD DOLK EVC FOXF1 GMPPA HPGD K ANSL1 ABAT ALDH3A2 ATP5A1 CCDC103 CLN5 CTSK DPAGT1 EVC2 FOXG1 GMPPB HPRT1 KAT6B ABCA12 ALDH4A1 ATP5E CCDC114 CLN6 CUBN DPM1 EXOC4 FOXH1 GNA11 HPSE2 KCNA2 ABCA3 ALDH5A1 ATP6AP2 CCDC151 CLN8 CUL4B DPM2 EXOSC3 FOXI1 GNAI3 HRAS KCNB1 ABCA4 ALDH7A1 ATP6V0A2 CCDC22 CLP1 CUL7 DPM3 EXPH5 FOXL2 GNAO1 HSD17B10 KCND2 ABCB11 ALDOA ATP6V1B1 CCDC39 CLPB CXCR4 DPP6 EYA1 FOXP1 GNAS HSD17B4 KCNE1 ABCB4 ALDOB ATP7A CCDC40 CLPP CYB5R3 DPYD EZH2 FOXP2 GNE HSD3B2 KCNE2 ABCB6 ALG1 ATP8A2 CCDC65 CNNM2 CYC1 DPYS F10 FOXP3 GNMT HSD3B7 KCNH2 ABCB7 ALG11 ATP8B1 CCDC78 CNTN1 CYP11B1 DRC1 F11 FOXRED1 GNPAT HSPD1 KCNH5 ABCC2 ALG12 ATPAF2 CCDC8 CNTNAP1 CYP11B2 DSC2 F13A1 FRAS1 GNPTAB HSPG2 KCNJ10 ABCC8 ALG13 ATR CCDC88C CNTNAP2 CYP17A1 DSG1 F13B FREM1 GNPTG HUWE1 KCNJ11 ABCC9 ALG14 ATRX CCND2 COA5 CYP1B1 DSP F2 FREM2 GNS HYDIN KCNJ13 ABCD3 ALG2 AUH CCNO COG1 CYP24A1 DST F5 FRMD7 GORAB HYLS1 KCNJ2 ABCD4 ALG3 B3GALNT2 CCS COG4 CYP26C1 DSTYK F7 FTCD GP1BA IBA57 KCNJ5 ABHD5 ALG6 B3GAT3 CCT5 COG5 CYP27A1 DTNA F8 FTO GP1BB ICK KCNJ8 ACAD8 ALG8 B3GLCT CD151 COG6 CYP27B1 DUOX2 F9 FUCA1 GP6 ICOS KCNK3 ACAD9 ALG9 -

Identification of Two Signaling Submodules Within the Crkii/ELMO
Cell Death and Differentiation (2007) 14, 963–972 & 2007 Nature Publishing Group All rights reserved 1350-9047/07 $30.00 www.nature.com/cdd Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells A-C Tosello-Trampont1,4, JM Kinchen1,2,4, E Brugnera1,3, LB Haney1, MO Hengartner2 and KS Ravichandran*,1 Removal of apoptotic cells is a dynamic process coordinated by ligands on apoptotic cells, and receptors and other signaling proteins on the phagocyte. One of the fundamental challenges is to understand how different phagocyte proteins form specific and functional complexes to orchestrate the recognition/removal of apoptotic cells. One evolutionarily conserved pathway involves the proteins cell death abnormal (CED)-2/chicken tumor virus no. 10 (CT10) regulator of kinase (Crk)II, CED-5/180 kDa protein downstream of chicken tumor virus no. 10 (Crk) (Dock180), CED-12/engulfment and migration (ELMO) and MIG-2/RhoG, leading to activation of the small GTPase CED-10/Rac and cytoskeletal remodeling to promote corpse uptake. Although the role of ELMO : Dock180 in regulating Rac activation has been well defined, the function of CED-2/CrkII in this complex is less well understood. Here, using functional studies in cell lines, we observe that a direct interaction between CrkII and Dock180 is not required for efficient removal of apoptotic cells. Similarly, mutants of CED-5 lacking the CED-2 interaction motifs could rescue engulfment and migration defects in CED-5 deficient worms. Mutants of CrkII and Dock180 that could not biochemically interact could colocalize in membrane ruffles. -

Disassembly of the Dying: Mechanisms and Functions
Review Disassembly of the Dying: Mechanisms and Functions 1 1, Georgia K. Atkin-Smith and Ivan K.H. Poon * The disassembly of an apoptotic cell into subcellular fragments, termed Trends apoptotic bodies (ApoBDs), is a hallmark of apoptosis. Although the genera- Apoptotic cell disassembly is a highly tion of ApoBDs is generally understood as being stochastic, it is becoming complex process regulated by a series of well-coordinated morphological increasingly clear that ApoBD formation is a highly regulated process involv- steps including apoptotic membrane ing distinct morphological steps and molecular factors. Functionally, ApoBDs blebbing, apoptotic protrusion forma- fi could facilitate the ef cient clearance of apoptotic material by surrounding tion, and fragmentation. phagocytes as well as mediate the transfer of biomolecules including micro- Plasma-membrane blebbing is not the RNAs and proteins between cells to aid in intercellular communications. sole process required for apoptotic Therefore, the formation of ApoBDs is an important process downstream body (ApoBD) formation, but mem- brane protrusions including microtu- from apoptotic cell death. We discuss here the mechanisms and functions bule spikes, apoptopodia, and of apoptotic cell disassembly. beaded apoptopodia may act in con- cert to aid the generation of ApoBDs. Cell Disassembly as a Key Downstream Process of Apoptotic Cell Death The mechanism of how apoptotic cells Billions of cells undergo apoptosis (a form of programmed cell death) daily as part of disassembly can determine the quantity and quality (size and contents) of physiological homeostasis [1]. At later stages of apoptosis, some cell types can generate ApoBDs. subcellular (1–5 mm) membrane-bound extracellular vesicles termed apoptotic bodies – (ApoBDs, see Glossary) [1 3]. -

Snapshot: Axon Guidance II Alex L
SnapShot: Axon Guidance II Alex L. Kolodkin1 and R. Jeroen Pasterkamp2 1Department of Neuroscience, HHMI, The Johns Hopkins University School of Medicine, Baltimore, MD 21212, USA 2Department of Neuroscience and Pharmacology, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, 3584 CG Utrecht, The Netherlands Axon guidance protein Chemotrophic effect Ligand-binding receptor Receptor processing Coreceptor Receptor signaling COFILIN, LIMK1, PI3K, BMP SE Repulsion BMP-RIB, BMP-RII - - SMAD1, SMAD6 DRAXIN SE Repulsion DCC - - - a2-CHIMAERIN, EPHEXIN-1, EPHRINA GPI Repulsion EPHA ADAM10 - NCK1/2, RAC1, RHOA, SPAR, VAV2/3 NCK1, PAK, p120GAP, RHOA, EPHRINB TM Repulsion EPHB MMP2/9 - ROCK, VAV2/3 TM Repulsion EPHRINA ADAM10 P75 FYN EPHA Attraction EPHRINA - RET - CDC42, DOCK180, GRB4, NCK2, EPHB TM Repulsion EPHRINB ADAM10, PS1 - PAK, RAC1 FGF SE Attraction FGFR1 - - - CDC42, DOCK180, ENA/VASP, APP, HSPG, ERK1/2, FAK, FYN, NCK1, N- SE Attraction DCC ADAM10, PS1 ROBO1 WASP, PAK, PI3K, PIP2, PKC, NETRIN RAC1, RHOA, TRIO, TRP Attraction NEOGENIN - - - Repulsion UNC5 - (DCC) FAK, SHP2, SRC FAK, LARG, LMO4, MYOIIA, RGM GPI Repulsion NEOGENIN TACE, PS1 UNC5 p120GAP, PKC, RAS, RHOA PLEXINA TM Repulsion SEMA1a - - PEBBLE, RHO, p190RHOGAP 14-3-3e, GYC76C, MICAL, SEMA1 TM Repulsion PLEXINA - OTK NERVY, PKA SE Repulsion PLEXINB - - RAC, RHO SEMA2 Attraction PLEXINB - - - AKT, COFILIN, CDK5, CRMP, FARP, NRP1/2, CAMs, SE Repulsion PLEXINA1-4 CALPAIN1 FYN, GSK3b, LIMK1, PI3K, RAC1, RTKs, ROBO SEMA3 RAP1, RAS, RND Attraction - - NRP1/2, -

Rho Guanine Nucleotide Exchange Factors: Regulators of Rho Gtpase Activity in Development and Disease
Oncogene (2014) 33, 4021–4035 & 2014 Macmillan Publishers Limited All rights reserved 0950-9232/14 www.nature.com/onc REVIEW Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease DR Cook1, KL Rossman2,3 and CJ Der1,2,3 The aberrant activity of Ras homologous (Rho) family small GTPases (20 human members) has been implicated in cancer and other human diseases. However, in contrast to the direct mutational activation of Ras found in cancer and developmental disorders, Rho GTPases are activated most commonly in disease by indirect mechanisms. One prevalent mechanism involves aberrant Rho activation via the deregulated expression and/or activity of Rho family guanine nucleotide exchange factors (RhoGEFs). RhoGEFs promote formation of the active GTP-bound state of Rho GTPases. The largest family of RhoGEFs is comprised of the Dbl family RhoGEFs with 70 human members. The multitude of RhoGEFs that activate a single Rho GTPase reflects the very specific role of each RhoGEF in controlling distinct signaling mechanisms involved in Rho activation. In this review, we summarize the role of Dbl RhoGEFs in development and disease, with a focus on Ect2 (epithelial cell transforming squence 2), Tiam1 (T-cell lymphoma invasion and metastasis 1), Vav and P-Rex1/2 (PtdIns(3,4,5)P3 (phosphatidylinositol (3,4,5)-triphosphate)-dependent Rac exchanger). Oncogene (2014) 33, 4021–4035; doi:10.1038/onc.2013.362; published online 16 September 2013 Keywords: Rac1; RhoA; Cdc42; guanine nucleotide exchange factors; cancer; -

Structural Analysis of Autoinhibition in the Ras-Specific Exchange Factor
RESEARCH ARTICLE elife.elifesciences.org Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1 Jeffrey S Iwig1,2, Yvonne Vercoulen3†, Rahul Das1,2†, Tiago Barros1,2,4, Andre Limnander3, Yan Che1,2, Jeffrey G Pelton2, David E Wemmer2,5,6, Jeroen P Roose3*, John Kuriyan1,2,4,5,6* 1Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, United States; 2California Institute for Quantitative Biosciences, University of California, Berkeley, Berkeley, United States; 3Department of Anatomy, University of California, San Francisco, San Francisco, United States; 4Howard Hughes Medical Institute, University of California, Berkeley, Berkeley, United States; 5Department of Chemistry, University of California, Berkeley, Berkeley, United States; 6Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, United States Abstract RasGRP1 and SOS are Ras-specific nucleotide exchange factors that have distinct roles in lymphocyte development. RasGRP1 is important in some cancers and autoimmune diseases but, in contrast to SOS, its regulatory mechanisms are poorly understood. Activating signals lead to the membrane recruitment of RasGRP1 and Ras engagement, but it is unclear how interactions between RasGRP1 and Ras are suppressed in the absence of such signals. We present a crystal structure of a fragment of RasGRP1 in which the Ras-binding site is blocked by an interdomain linker and the membrane-interaction surface of RasGRP1 is hidden within a dimerization interface that may be stabilized by the C-terminal oligomerization domain. NMR data demonstrate that calcium binding to the regulatory module generates substantial conformational changes that are incompatible with the inactive assembly. These features allow RasGRP1 to be maintained in an inactive state that is poised for activation by calcium and membrane-localization signals.