Receptor Internalization Assays
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes
Tohoku J. Exp. Med., 2011,Differential 223, 161-176 Gene Expression in OPCs, Oligodendrocytes and Type II Astrocytes 161 Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes Jian-Guo Hu,1,2,* Yan-Xia Wang,3,* Jian-Sheng Zhou,2 Chang-Jie Chen,4 Feng-Chao Wang,1 Xing-Wu Li1 and He-Zuo Lü1,2 1Department of Clinical Laboratory Science, The First Affiliated Hospital of Bengbu Medical College, Bengbu, P.R. China 2Anhui Key Laboratory of Tissue Transplantation, Bengbu Medical College, Bengbu, P.R. China 3Department of Neurobiology, Shanghai Jiaotong University School of Medicine, Shanghai, P.R. China 4Department of Laboratory Medicine, Bengbu Medical College, Bengbu, P.R. China Oligodendrocyte precursor cells (OPCs) are bipotential progenitor cells that can differentiate into myelin-forming oligodendrocytes or functionally undetermined type II astrocytes. Transplantation of OPCs is an attractive therapy for demyelinating diseases. However, due to their bipotential differentiation potential, the majority of OPCs differentiate into astrocytes at transplanted sites. It is therefore important to understand the molecular mechanisms that regulate the transition from OPCs to oligodendrocytes or astrocytes. In this study, we isolated OPCs from the spinal cords of rat embryos (16 days old) and induced them to differentiate into oligodendrocytes or type II astrocytes in the absence or presence of 10% fetal bovine serum, respectively. RNAs were extracted from each cell population and hybridized to GeneChip with 28,700 rat genes. Using the criterion of fold change > 4 in the expression level, we identified 83 genes that were up-regulated and 89 genes that were down-regulated in oligodendrocytes, and 92 genes that were up-regulated and 86 that were down-regulated in type II astrocytes compared with OPCs. -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Immunobiology of VPAC2 Receptor
Aus der Medizinischen Poliklinik-Innenstadt der Ludwig-Maximilians-Universität München Direktor: Professor Dr. med. M. Reincke Immunobiology of the VPAC2 Receptor Dissertation zum Erwerb des Doktorgrades der Medizin an der Medizinischen Fakultät der Ludwig-Maximilians-Universität zu München Vorgelegt von Carola Grinninger aus München 2010 Mit Genehmigung der Medizinischen Fakultät der Universität München Berichterstatter: Prof. Dr. med. S. Schewe Mitberichterstatter: PD Dr. B. Bachmeier Prof. Dr. H. Kellner Prof. Dr. U. Gresser Betreuung durch den promovierten Mitarbeiter: PD Dr. P. J. Nelson Dekan: Prof. Dr. med. Dr. h.c. M. Reiser, FACR, FRCR Tag der mündlichen Prüfung: 07.10.2010 2 CONTENTS Contents……………………………………………………………………….…3 Summary…………………………………………………………………………5 1. Introduction……………………………………………………………………10 1.1 General introduction……………………………………………………...10 1.2 G-Protein-coupled Receptors…………………………………………….10 1.3 G-protein-coupled signalling…………………………………………….13 1.4 Molecularbiology and structure of VPAC Receptors………………...….15 1.5 Vasoactive Intestinal Peptide (VIP)……………………………………...22 1.5.1 Biochemical structure of vasoactive intestinal peptide………...............22 1.6 VIP and intracellular signaling…………………………………………...25 1.7 VIP and rheumatoid arthritis…………………………………..…………26 1.8 Hypothesis………………………………………………………………..29 2. Materials……………………………………………………..………………31 2.1 Bacteria ……………………………………………………………….…31 2.2 Vectors……………………………………………………………….…..31 2.3 Cells………………………………………………………………….…..32 2.4 Mice……………………………………………………………….….….33 2.5 Oligonucleotides………………………………………………….….…..33 -

Current Topics in Medicinal Chemistry, 2019, 19, 1399-1417 REVIEW ARTICLE
Send Orders for Reprints to [email protected] 1399 Current Topics in Medicinal Chemistry, 2019, 19, 1399-1417 REVIEW ARTICLE ISSN: 1568-0266 eISSN: 1873-4294 Impact Factor: Targeting the PAC1 Receptor for Neurological and Metabolic Disorders 3.442 The international journal for in-depth reviews on Current Topics in Medicinal Chemistry BENTHAM SCIENCE Chenyi Liao1, Mathilde P. de Molliens2, Severin T. Schneebeli1, Matthias Brewer1, Gaojie Song3, David Chatenet2, Karen M. Braas4, Victor May4,* and Jianing Li1,* 1Department of Chemistry, University of Vermont, Burlington, VT 05405, USA; 2INRS – Institut Armand-Frappier, 531 boul. des Prairies, Laval, QC H7V 1B7, Canada; 3Shanghai Key Laboratory of Regulatory Biology, Institute of Bio- medical Sciences and School of Life Sciences, East China Normal University, Shanghai, 200241, P.R. China; 4Department of Neurological Sciences, University of Vermont, Larner College of Medicine, 149 Beaumont Avenue, Bur- lington, VT 05405, USA Abstract: The pituitary adenylate cyclase-activating polypeptide (PACAP)-selective PAC1 receptor (PAC1R, ADCYAP1R1) is a member of the vasoactive intestinal peptide (VIP)/secretin/glucagon fam- ily of G protein-coupled receptors (GPCRs). PAC1R has been shown to play crucial roles in the central and peripheral nervous systems. The activation of PAC1R initiates diverse downstream signal transduc- tion pathways, including adenylyl cyclase, phospholipase C, MEK/ERK, and Akt pathways that regu- late a number of physiological systems to maintain functional homeostasis. Accordingly, at times of tissue injury or insult, PACAP/PAC1R activation of these pathways can be trophic to blunt or delay apoptotic events and enhance cell survival. Enhancing PAC1R signaling under these conditions has the potential to mitigate cellular damages associated with cerebrovascular trauma (including stroke), neu- A R T I C L E H I S T O R Y rodegeneration (such as Parkinson’s and Alzheimer's disease), or peripheral organ insults. -

Neuronal, Stromal, and T-Regulatory Cell Crosstalk in Murine Skeletal Muscle
Neuronal, stromal, and T-regulatory cell crosstalk in murine skeletal muscle Kathy Wanga,b,1,2, Omar K. Yaghia,b,1, Raul German Spallanzania,b,1, Xin Chena,b,3, David Zemmoura,b,4, Nicole Laia, Isaac M. Chiua, Christophe Benoista,b,5, and Diane Mathisa,b,5 aDepartment of Immunology, Harvard Medical School, Boston, MA 02115; and bEvergrande Center for Immunologic Diseases, Harvard Medical School and Brigham and Women’s Hospital, Boston, MA 02115 Contributed by Diane Mathis, January 15, 2020 (sent for review December 23, 2019; reviewed by David A. Hafler and Jeffrey V. Ravetch) A distinct population of Foxp3+CD4+ regulatory T (Treg) cells pro- reduced in aged mice characterized by poor muscle regeneration + motes repair of acutely or chronically injured skeletal muscle. The (7). IL-33 mSCs can be found in close association with nerve accumulation of these cells depends critically on interleukin (IL)-33 pro- structures in skeletal muscle, including nerve fibers, nerve bun- duced by local mesenchymal stromal cells (mSCs). An intriguing phys- + dles, and muscle spindles that control proprioception (7). ical association among muscle nerves, IL-33 mSCs, and Tregs has been Given the intriguing functional and/or physical associations reported, and invites a deeper exploration of this cell triumvirate. Here + among muscle nerves, mSCs, and Tregs, and in particular, their we evidence a striking proximity between IL-33 muscle mSCs and co-ties to IL-33, we were inspired to more deeply explore this both large-fiber nerve bundles and small-fiber sensory neurons; report axis. Here, we used whole-mount immunohistochemical imag- that muscle mSCs transcribe an array of genes encoding neuropep- ing as well as population-level and single-cell RNA sequencing tides, neuropeptide receptors, and other nerve-related proteins; define (scRNA-seq) to examine the neuron/mSC/Treg triumvirate in muscle mSC subtypes that express both IL-33 and the receptor for the calcitonin-gene–related peptide (CGRP); and demonstrate that up- or hindlimb muscles. -

GPCR Expression Profiles Were Determined Using
Supplemental Figures and Tables for Tischner et al., 2017 Supplemental Figure 1: GPCR expression profiles were determined using the NanoString nCounter System in 250 ng of pooled cell RNA obtained from freshly isolated CD4 T cells from naïve lymph nodes (CD4ln), spinal cord infiltrating CD4 T cells at peak EAE disease (CD4sc), and primary lung endothelial cells (luEC). Supplemental Figure 2: Array design and quality controls. A, Sorted leukocytes or endothelial cells were subjected to single‐cell expression analysis and re‐evaluated based on the expression of various identity‐defining genes. B, Expression of identity‐defining and quality control genes after deletion of contaminating or reference gene‐negative cells. Expression data are calculated as 2(Limit of detection(LoD) Ct – sample Ct) ; LoD Ct was set to 24. Supplemental Figure 3: Overview over GPCR expression frequencies in different freshly isolated immune cell populations and spinal cord endothelial cells as determined by single cell RT‐PCR. Abbreviations: CD4ln‐Tcon/CD4ln‐Treg, conventional (con) and regulatory (reg) CD4 T cells from lymph nodes (CD4ln) of naïve mice; CD4dr/CD4sc, CD4 T cells from draining lymph nodes (dr) or spinal cord (sc) at peak EAE disease; CD4spn2D/ CD4spn2DTh1/ CD4spn2DTh17, splenic CD4 T cells from 2D2 T cell receptor transgenic mice before (2D) and after in vitro differentiation towards Th1 (2DTh1) or Th17 (2DTh17); MonoSpn, splenic monocytes; CD11b_sc, spinal cord infiltrating CD11b‐ positive cells; sc_microglia, Ccr2neg,Cx3cr1pos microglia from spinal cord at peak disease; sc_macrophages, CCr2pos;Cx3cr1lo/neg macrophages from spinal cord at peak disease; BMDM_M1/BMDM_M2, bone marrow‐derived macrophages differentiated towards M1 or M2; ECscN and ECscEAE, spinal cord endothelial cells from naïve mice (N) and at peak EAE disease (EAE); SMC, smooth muscle cells from various vessel types (included as positive control to ascertain primer functionality). -
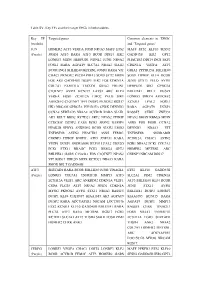
Targeted Genes Common Elements In
Table SV. Key TFs and their target DEGs in hub modules. Key TF Targeted genes Common elements in ‘DEGs’ (module) and ‘Targeted genes’ JUN HOMER2 ATF3 VEGFA FOSB NR4A3 MAFF ETS2 MAFF ETS2 KLF10 SESN2 (Purple) JOSD1 ATF3 RARA ATF3 BCOR DDIT4 IER2 GADD45B IER2 GPT2 LONRF3 MIDN HERPUD1 NDNL2 JUNB NR4A2 PHACTR3 DDIT4 ING1 SKP2 FOSL1 RARA AGPAT9 SLC7A1 NR4A2 SIAH2 CDKN1A VEGFA ATF3 BCOR ING1 BHLHE40 METRNL JOSD1 RARA VIT GRIA2 PPP1R15A BHLHE40 CHAC1 PKNOX2 PLCB4 PHF13 SOX9 SYT2 MIDN SOX9 ITPRIP KLF4 BCOR FOS AK5 GADD45B DUSP1 IER2 FOS CDKN1A JUNB STX11 PELO AVPI1 COL7A1 FAM131A TBCCD1 GRIA2 ERLIN1 HERPUD1 SIK1 GPRC5A C1QTNF7 AVPI1 KCNJ15 LATS2 ARC KLF4 BHLHE41 RELT DUSP2 VEGFA FOSB ZC3H12A LMO2 PELO SIK1 LONRF3 SPRY4 ARHGEF2 ARHGEF2 C1QTNF7 TFPI DUSP2 PKNOX2 RGS17 KCNJ15 LPAL2 FOSL1 HK1 NRCAM GPRC5A PPP1R15A CERK DENND3 RARA AGPAT9 DUSP1 CCNA2 SERTAD3 NR4A2 ACVR1B RARA SIAH1 RASSF5 CERK ZNF331 AK5 RELT BRD2 KCTD21 SKP2 NPAS2 ITPRIP NPAS2 SMOX RBM24 MIDN CCDC85C DUSP2 CARS EGR1 SESN2 RASSF5 ASNS FOS FOSB CCNA2 PIP4K2B SPRY4 ANKRD52 BCOR SIAH2 LMO2 DENND3 NR4A3 VIT TNFRSF1B ASTN2 PHACTR3 ASNS FEM1C TNFRSF1B SNORA80B CSRNP3 ITPRIP BNIP3L ATF3 ZNF331 RARA ZC3H12A CHAC1 ASTN2 VEZF1 DUSP1 SNORA80B KLF10 LPAL2 UBE2O EGR1 NR4A2 PCK1 COL7A1 PCK1 STX11 RRAGC PCK1 RBM24 GPT2 HOMER2 METRNL ARC BHLHE41 GARS C15orf41 FOS C1QTNF7 NPAS2 CSRNP3 NRCAM RGS17 VIT RGS17 UBE2O SOX9 KCTD21 NR4A3 RARA SMOX RELT GADD45B ATF3 SERTAD1 RARA BCOR BHLHE40 JUNB TINAGL1 ETS2 KLF10 GADD45B (Purple) LONRF3 COL7A1 TNFRSF1B MMP13 ATF3 SLC2A1 PIM2 CDKN1A ZC3H12A VEZF1 ARC ANKRD52 -

Robles JTO Supplemental Digital Content 1
Supplementary Materials An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma based on mRNA, microRNA and DNA Methylation Biomarkers Ana I. Robles1, Eri Arai2, Ewy A. Mathé1, Hirokazu Okayama1, Aaron Schetter1, Derek Brown1, David Petersen3, Elise D. Bowman1, Rintaro Noro1, Judith A. Welsh1, Daniel C. Edelman3, Holly S. Stevenson3, Yonghong Wang3, Naoto Tsuchiya4, Takashi Kohno4, Vidar Skaug5, Steen Mollerup5, Aage Haugen5, Paul S. Meltzer3, Jun Yokota6, Yae Kanai2 and Curtis C. Harris1 Affiliations: 1Laboratory of Human Carcinogenesis, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 2Division of Molecular Pathology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 3Genetics Branch, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 4Division of Genome Biology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 5Department of Chemical and Biological Working Environment, National Institute of Occupational Health, NO-0033 Oslo, Norway. 6Genomics and Epigenomics of Cancer Prediction Program, Institute of Predictive and Personalized Medicine of Cancer (IMPPC), 08916 Badalona (Barcelona), Spain. List of Supplementary Materials Supplementary Materials and Methods Fig. S1. Hierarchical clustering of based on CpG sites differentially-methylated in Stage I ADC compared to non-tumor adjacent tissues. Fig. S2. Confirmatory pyrosequencing analysis of DNA methylation at the HOXA9 locus in Stage I ADC from a subset of the NCI microarray cohort. 1 Fig. S3. Methylation Beta-values for HOXA9 probe cg26521404 in Stage I ADC samples from Japan. Fig. S4. Kaplan-Meier analysis of HOXA9 promoter methylation in a published cohort of Stage I lung ADC (J Clin Oncol 2013;31(32):4140-7). Fig. S5. Kaplan-Meier analysis of a combined prognostic biomarker in Stage I lung ADC. -

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Peripheral Nerve Single-Cell Analysis Identifies Mesenchymal Ligands That Promote Axonal Growth
Research Article: New Research Development Peripheral Nerve Single-Cell Analysis Identifies Mesenchymal Ligands that Promote Axonal Growth Jeremy S. Toma,1 Konstantina Karamboulas,1,ª Matthew J. Carr,1,2,ª Adelaida Kolaj,1,3 Scott A. Yuzwa,1 Neemat Mahmud,1,3 Mekayla A. Storer,1 David R. Kaplan,1,2,4 and Freda D. Miller1,2,3,4 https://doi.org/10.1523/ENEURO.0066-20.2020 1Program in Neurosciences and Mental Health, Hospital for Sick Children, 555 University Avenue, Toronto, Ontario M5G 1X8, Canada, 2Institute of Medical Sciences University of Toronto, Toronto, Ontario M5G 1A8, Canada, 3Department of Physiology, University of Toronto, Toronto, Ontario M5G 1A8, Canada, and 4Department of Molecular Genetics, University of Toronto, Toronto, Ontario M5G 1A8, Canada Abstract Peripheral nerves provide a supportive growth environment for developing and regenerating axons and are es- sential for maintenance and repair of many non-neural tissues. This capacity has largely been ascribed to paracrine factors secreted by nerve-resident Schwann cells. Here, we used single-cell transcriptional profiling to identify ligands made by different injured rodent nerve cell types and have combined this with cell-surface mass spectrometry to computationally model potential paracrine interactions with peripheral neurons. These analyses show that peripheral nerves make many ligands predicted to act on peripheral and CNS neurons, in- cluding known and previously uncharacterized ligands. While Schwann cells are an important ligand source within injured nerves, more than half of the predicted ligands are made by nerve-resident mesenchymal cells, including the endoneurial cells most closely associated with peripheral axons. At least three of these mesen- chymal ligands, ANGPT1, CCL11, and VEGFC, promote growth when locally applied on sympathetic axons. -

RNA-Seq Data from 53 Tissues Provided by 544 Donors, with a Total Of
1 Supplementary Materials 2 3 Supplementary Methods 4 Co-expression analysis: RNA-seq data from 53 tissues provided by 544 donors, with a total of 5 8555 samples, were downloaded from gtexportal.org (GTEx_Analysis_v6p_RNA-seq_RNA- 6 SeQCv1.1.8_gene_rpkm.gct.gz) (41). Downloaded data was provided as reads per kilobase per 7 million mapped reads (RPKM). Only samples passing quality control were included in the 8 dataset. Read counts and RPKM values were produced with RNA-SeqC; importantly, reads were 9 mapped to a single gene (see gtexportal.org documentation for more information). For quantile 10 normalization, gene expression across an individual sample was fit to the averaged distribution 11 observed across samples. Prior to implementing spearman correlation analysis, the median 12 normalized expression per tissue was calculated to account for differences in the number of 13 samples per tissue type. Spearman correlation coefficient was calculated for each RAMP/non- 14 olfactory GPCR pair. GPCR clusters were assigned as per Fredriksson et al (42). 15 16 Statistical comparison of data sets: The data obtained using anti-GPCR capture Abs was compared 17 to the data obtained using the epitope-tag capture methods. Based upon the data set for GPCR- 18 RAMP complexes derived from the epitope-tag capture Abs, a matrix of hypothetical outcomes for 19 the data set derived from the anti-GPCR Ab capture strategy was constructed. To compare the two 20 matrices, the results were converted to binary score matrices (0,1) and the two matrices (epitope- 21 tag Ab capture versus direct Ab capture). A Z-score threshold of 1.645 was applied for the anti- 22 GPCR Ab data set of 1.645, which corresponds to a confidence interval of 95% for a single-tailed 23 test (fig S5).