Immunobiology of VPAC2 Receptor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -
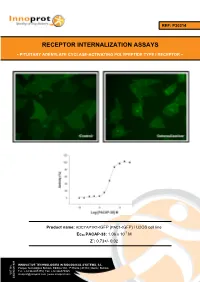
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes
Tohoku J. Exp. Med., 2011,Differential 223, 161-176 Gene Expression in OPCs, Oligodendrocytes and Type II Astrocytes 161 Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes Jian-Guo Hu,1,2,* Yan-Xia Wang,3,* Jian-Sheng Zhou,2 Chang-Jie Chen,4 Feng-Chao Wang,1 Xing-Wu Li1 and He-Zuo Lü1,2 1Department of Clinical Laboratory Science, The First Affiliated Hospital of Bengbu Medical College, Bengbu, P.R. China 2Anhui Key Laboratory of Tissue Transplantation, Bengbu Medical College, Bengbu, P.R. China 3Department of Neurobiology, Shanghai Jiaotong University School of Medicine, Shanghai, P.R. China 4Department of Laboratory Medicine, Bengbu Medical College, Bengbu, P.R. China Oligodendrocyte precursor cells (OPCs) are bipotential progenitor cells that can differentiate into myelin-forming oligodendrocytes or functionally undetermined type II astrocytes. Transplantation of OPCs is an attractive therapy for demyelinating diseases. However, due to their bipotential differentiation potential, the majority of OPCs differentiate into astrocytes at transplanted sites. It is therefore important to understand the molecular mechanisms that regulate the transition from OPCs to oligodendrocytes or astrocytes. In this study, we isolated OPCs from the spinal cords of rat embryos (16 days old) and induced them to differentiate into oligodendrocytes or type II astrocytes in the absence or presence of 10% fetal bovine serum, respectively. RNAs were extracted from each cell population and hybridized to GeneChip with 28,700 rat genes. Using the criterion of fold change > 4 in the expression level, we identified 83 genes that were up-regulated and 89 genes that were down-regulated in oligodendrocytes, and 92 genes that were up-regulated and 86 that were down-regulated in type II astrocytes compared with OPCs. -

MONOCLONAL ANTI-VASOACTIVE INTESTINAL PEPTIDE RECEPTOR 1 (VIPR 1, VPAC1) Clone AS58 Purified Mouse Immunoglobulin
MONOCLONAL ANTI-VASOACTIVE INTESTINAL PEPTIDE RECEPTOR 1 (VIPR 1, VPAC1) Clone AS58 Purified Mouse Immunoglobulin Product Number V1631 Product Description Although structurally related, VIPR1 and VIPR2 exhibit Monoclonal Anti-Vasoactive Intestinal Peptide differences in expression and function and VIP has a Receptor 1 (VIPR1) (mouse IgG2a isotype) is derived 3-10 fold preference for VIPR1 over VIPR2 receptors. from the hybridoma produced by the fusion of mouse myeloma cells and splenocytes from a BALB/c mouse VIPR1 is expressed throughout the central nervous immunized with a unique peptide corresponding to a system (predominantly in the cerebral cortex and portion of human Vasoactive Intestinal Peptide hippocampus), in peripheral tissues including liver, lung Receptor 1 (VIPR1). The antibody was purified from and intestine and in T lymphocytes.14 VIPR1 mediates tissue culture supernatant using immobilized Protein G. suppression of chemotaxis and matrix metalloprotein- ase expression elicited by some cytokines and chemo- Monoclonal Anti-Vasoactive Intestinal Peptide kines, tumor cell migration induced by VIP, and vaso- Receptor 1 (VIPR1) recognizes VIPR1 protein from dilation. human and rat tissue by immunoblotting and by flow cytometric analysis of human cells using indirect VIPR2 is expressed throughout the central nervous immunofluorescence. The antibody does not recognize system, but to varying degrees. The highest expression VIPR2 protein. levels are in the thalamus and suprachiasmatic nucleus, but VIPR2 is also present in the hippocampus, -

Mouse Genotypes Drive the Liver and Adrenal Gland Clocks
www.nature.com/scientificreports OPEN Mouse genotypes drive the liver and adrenal gland clocks Rok Košir1,*, Uršula Prosenc Zmrzljak1,*, Anja Korenčič1, Peter Juvan1, Jure Ačimovič2 & Damjana Rozman1,2 Received: 15 November 2015 Circadian rhythms regulate a plethora of physiological processes. Perturbations of the rhythm can Accepted: 25 July 2016 result in pathologies which are frequently studied in inbred mouse strains. We show that the genotype Published: 18 August 2016 of mouse lines defines the circadian gene expression patterns. Expression of majority of core clock and output metabolic genes are phase delayed in the C56BL/6J line compared to 129S2 in the adrenal glands and the liver. Circadian amplitudes are generally higher in the 129S2 line. Experiments in dark – dark (DD) and light – dark conditions (LD), exome sequencing and data mining proposed that mouse lines differ in single nucleotide variants in the binding regions of clock related transcription factors in open chromatin regions. A possible mechanisms of differential circadian expression could be the entrainment and transmission of the light signal to peripheral organs. This is supported by the genotype effect in adrenal glands that is largest under LD, and by the high number of single nucleotide variants in the Receptor, Kinase and G-protein coupled receptor Panther molecular function categories. Different phenotypes of the two mouse lines and changed amino acid sequence of the Period 2 protein possibly contribute further to the observed differences in circadian gene expression. The majority of organisms on Earth have evolved a robust inner body circadian (circa = approximately, dian = day) clock that ticks away in most of their cells. -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Multi-Functionality of Proteins Involved in GPCR and G Protein Signaling: Making Sense of Structure–Function Continuum with In
Cellular and Molecular Life Sciences (2019) 76:4461–4492 https://doi.org/10.1007/s00018-019-03276-1 Cellular andMolecular Life Sciences REVIEW Multi‑functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder‑based proteoforms Alexander V. Fonin1 · April L. Darling2 · Irina M. Kuznetsova1 · Konstantin K. Turoverov1,3 · Vladimir N. Uversky2,4 Received: 5 August 2019 / Revised: 5 August 2019 / Accepted: 12 August 2019 / Published online: 19 August 2019 © Springer Nature Switzerland AG 2019 Abstract GPCR–G protein signaling system recognizes a multitude of extracellular ligands and triggers a variety of intracellular signal- ing cascades in response. In humans, this system includes more than 800 various GPCRs and a large set of heterotrimeric G proteins. Complexity of this system goes far beyond a multitude of pair-wise ligand–GPCR and GPCR–G protein interactions. In fact, one GPCR can recognize more than one extracellular signal and interact with more than one G protein. Furthermore, one ligand can activate more than one GPCR, and multiple GPCRs can couple to the same G protein. This defnes an intricate multifunctionality of this important signaling system. Here, we show that the multifunctionality of GPCR–G protein system represents an illustrative example of the protein structure–function continuum, where structures of the involved proteins represent a complex mosaic of diferently folded regions (foldons, non-foldons, unfoldons, semi-foldons, and inducible foldons). The functionality of resulting highly dynamic conformational ensembles is fne-tuned by various post-translational modifcations and alternative splicing, and such ensembles can undergo dramatic changes at interaction with their specifc partners. -

1 Supplemental Material Maresin 1 Activates LGR6 Receptor
Supplemental Material Maresin 1 Activates LGR6 Receptor Promoting Phagocyte Immunoresolvent Functions Nan Chiang, Stephania Libreros, Paul C. Norris, Xavier de la Rosa, Charles N. Serhan Center for Experimental Therapeutics and Reperfusion Injury, Department of Anesthesiology, Perioperative and Pain Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts 02115, USA. 1 Supplemental Table 1. Screening of orphan GPCRs with MaR1 Vehicle Vehicle MaR1 MaR1 mean RLU > GPCR ID SD % Activity Mean RLU Mean RLU + 2 SD Mean RLU Vehicle mean RLU+2 SD? ADMR 930920 33283 997486.5381 863760 -7% BAI1 172580 18362 209304.1828 176160 2% BAI2 26390 1354 29097.71737 26240 -1% BAI3 18040 758 19555.07976 18460 2% CCRL2 15090 402 15893.6583 13840 -8% CMKLR2 30080 1744 33568.954 28240 -6% DARC 119110 4817 128743.8016 126260 6% EBI2 101200 6004 113207.8197 105640 4% GHSR1B 3940 203 4345.298244 3700 -6% GPR101 41740 1593 44926.97349 41580 0% GPR103 21413 1484 24381.25067 23920 12% NO GPR107 366800 11007 388814.4922 360020 -2% GPR12 77980 1563 81105.4653 76260 -2% GPR123 1485190 46446 1578081.986 1342640 -10% GPR132 860940 17473 895885.901 826560 -4% GPR135 18720 1656 22032.6827 17540 -6% GPR137 40973 2285 45544.0809 39140 -4% GPR139 438280 16736 471751.0542 413120 -6% GPR141 30180 2080 34339.2307 29020 -4% GPR142 105250 12089 129427.069 101020 -4% GPR143 89390 5260 99910.40557 89380 0% GPR146 16860 551 17961.75617 16240 -4% GPR148 6160 484 7128.848113 7520 22% YES GPR149 50140 934 52008.76073 49720 -1% GPR15 10110 1086 12282.67884 -

Current Topics in Medicinal Chemistry, 2019, 19, 1399-1417 REVIEW ARTICLE
Send Orders for Reprints to [email protected] 1399 Current Topics in Medicinal Chemistry, 2019, 19, 1399-1417 REVIEW ARTICLE ISSN: 1568-0266 eISSN: 1873-4294 Impact Factor: Targeting the PAC1 Receptor for Neurological and Metabolic Disorders 3.442 The international journal for in-depth reviews on Current Topics in Medicinal Chemistry BENTHAM SCIENCE Chenyi Liao1, Mathilde P. de Molliens2, Severin T. Schneebeli1, Matthias Brewer1, Gaojie Song3, David Chatenet2, Karen M. Braas4, Victor May4,* and Jianing Li1,* 1Department of Chemistry, University of Vermont, Burlington, VT 05405, USA; 2INRS – Institut Armand-Frappier, 531 boul. des Prairies, Laval, QC H7V 1B7, Canada; 3Shanghai Key Laboratory of Regulatory Biology, Institute of Bio- medical Sciences and School of Life Sciences, East China Normal University, Shanghai, 200241, P.R. China; 4Department of Neurological Sciences, University of Vermont, Larner College of Medicine, 149 Beaumont Avenue, Bur- lington, VT 05405, USA Abstract: The pituitary adenylate cyclase-activating polypeptide (PACAP)-selective PAC1 receptor (PAC1R, ADCYAP1R1) is a member of the vasoactive intestinal peptide (VIP)/secretin/glucagon fam- ily of G protein-coupled receptors (GPCRs). PAC1R has been shown to play crucial roles in the central and peripheral nervous systems. The activation of PAC1R initiates diverse downstream signal transduc- tion pathways, including adenylyl cyclase, phospholipase C, MEK/ERK, and Akt pathways that regu- late a number of physiological systems to maintain functional homeostasis. Accordingly, at times of tissue injury or insult, PACAP/PAC1R activation of these pathways can be trophic to blunt or delay apoptotic events and enhance cell survival. Enhancing PAC1R signaling under these conditions has the potential to mitigate cellular damages associated with cerebrovascular trauma (including stroke), neu- A R T I C L E H I S T O R Y rodegeneration (such as Parkinson’s and Alzheimer's disease), or peripheral organ insults. -

ADRB2-Targeting Therapies for Prostate Cancer
cancers Review ADRB2-Targeting Therapies for Prostate Cancer George Kulik 1,2 1 Department of Cancer Biology, Wake Forest University Health Sciences, Medical Center Blvd, Winston-Salem, NC 27157, USA; [email protected] or [email protected] 2 Department of Life Sciences, Alfaisal University, Riyadh 11533, Saudi Arabia Received: 18 February 2019; Accepted: 8 March 2019; Published: 13 March 2019 Abstract: There is accumulating evidence that β-2 adrenergic receptor (ADRB2) signaling contributes to the progression and therapy resistance of prostate cancer, whereas availability of clinically tested β-blocker propranolol makes this pathway especially attractive as potential therapeutic target. Yet even in tumors with active ADRB2 signaling propranolol may be ineffective. Inhibition of apoptosis is one of the major mechanisms by which activation of ADRB2 contributes to prostate cancer pathophysiology. The signaling network that controls apoptosis in prostate tumors is highly redundant, with several signaling pathways targeting a few critical apoptosis regulatory molecules. Therefore, a comprehensive analysis of ADRB2 signaling in the context of other signaling mechanisms is necessary to identify patients who will benefit from propranolol therapy. This review discusses how information on the antiapoptotic mechanisms activated by ADRB2 can guide clinical trials of ADRB2 antagonist propranolol as potential life-extending therapy for prostate cancer. To select patients for clinical trials of propranolol three classes of biomarkers are proposed. First, biomarkers of ADRB2/cAMP-dependent protein kinase (PKA) pathway activation; second, biomarkers that inform about activation of other signaling pathways unrelated to ADRB2; third, apoptosis regulatory molecules controlled by ADRB2 signaling and other survival signaling pathways. Keywords: β-2 adrenergic receptor (ADRB2); cAMP-dependent protein kinase (PKA); prostate cancer; propranolol; apoptosis; clinical trial; BCL-2-associated death promoter (BAD); myeloid cell leukemia 1 (MCL-1) 1. -

Neuronal, Stromal, and T-Regulatory Cell Crosstalk in Murine Skeletal Muscle
Neuronal, stromal, and T-regulatory cell crosstalk in murine skeletal muscle Kathy Wanga,b,1,2, Omar K. Yaghia,b,1, Raul German Spallanzania,b,1, Xin Chena,b,3, David Zemmoura,b,4, Nicole Laia, Isaac M. Chiua, Christophe Benoista,b,5, and Diane Mathisa,b,5 aDepartment of Immunology, Harvard Medical School, Boston, MA 02115; and bEvergrande Center for Immunologic Diseases, Harvard Medical School and Brigham and Women’s Hospital, Boston, MA 02115 Contributed by Diane Mathis, January 15, 2020 (sent for review December 23, 2019; reviewed by David A. Hafler and Jeffrey V. Ravetch) A distinct population of Foxp3+CD4+ regulatory T (Treg) cells pro- reduced in aged mice characterized by poor muscle regeneration + motes repair of acutely or chronically injured skeletal muscle. The (7). IL-33 mSCs can be found in close association with nerve accumulation of these cells depends critically on interleukin (IL)-33 pro- structures in skeletal muscle, including nerve fibers, nerve bun- duced by local mesenchymal stromal cells (mSCs). An intriguing phys- + dles, and muscle spindles that control proprioception (7). ical association among muscle nerves, IL-33 mSCs, and Tregs has been Given the intriguing functional and/or physical associations reported, and invites a deeper exploration of this cell triumvirate. Here + among muscle nerves, mSCs, and Tregs, and in particular, their we evidence a striking proximity between IL-33 muscle mSCs and co-ties to IL-33, we were inspired to more deeply explore this both large-fiber nerve bundles and small-fiber sensory neurons; report axis. Here, we used whole-mount immunohistochemical imag- that muscle mSCs transcribe an array of genes encoding neuropep- ing as well as population-level and single-cell RNA sequencing tides, neuropeptide receptors, and other nerve-related proteins; define (scRNA-seq) to examine the neuron/mSC/Treg triumvirate in muscle mSC subtypes that express both IL-33 and the receptor for the calcitonin-gene–related peptide (CGRP); and demonstrate that up- or hindlimb muscles. -

GPCR Expression Profiles Were Determined Using
Supplemental Figures and Tables for Tischner et al., 2017 Supplemental Figure 1: GPCR expression profiles were determined using the NanoString nCounter System in 250 ng of pooled cell RNA obtained from freshly isolated CD4 T cells from naïve lymph nodes (CD4ln), spinal cord infiltrating CD4 T cells at peak EAE disease (CD4sc), and primary lung endothelial cells (luEC). Supplemental Figure 2: Array design and quality controls. A, Sorted leukocytes or endothelial cells were subjected to single‐cell expression analysis and re‐evaluated based on the expression of various identity‐defining genes. B, Expression of identity‐defining and quality control genes after deletion of contaminating or reference gene‐negative cells. Expression data are calculated as 2(Limit of detection(LoD) Ct – sample Ct) ; LoD Ct was set to 24. Supplemental Figure 3: Overview over GPCR expression frequencies in different freshly isolated immune cell populations and spinal cord endothelial cells as determined by single cell RT‐PCR. Abbreviations: CD4ln‐Tcon/CD4ln‐Treg, conventional (con) and regulatory (reg) CD4 T cells from lymph nodes (CD4ln) of naïve mice; CD4dr/CD4sc, CD4 T cells from draining lymph nodes (dr) or spinal cord (sc) at peak EAE disease; CD4spn2D/ CD4spn2DTh1/ CD4spn2DTh17, splenic CD4 T cells from 2D2 T cell receptor transgenic mice before (2D) and after in vitro differentiation towards Th1 (2DTh1) or Th17 (2DTh17); MonoSpn, splenic monocytes; CD11b_sc, spinal cord infiltrating CD11b‐ positive cells; sc_microglia, Ccr2neg,Cx3cr1pos microglia from spinal cord at peak disease; sc_macrophages, CCr2pos;Cx3cr1lo/neg macrophages from spinal cord at peak disease; BMDM_M1/BMDM_M2, bone marrow‐derived macrophages differentiated towards M1 or M2; ECscN and ECscEAE, spinal cord endothelial cells from naïve mice (N) and at peak EAE disease (EAE); SMC, smooth muscle cells from various vessel types (included as positive control to ascertain primer functionality).