Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Strategies to Increase ß-Cell Mass Expansion
This electronic thesis or dissertation has been downloaded from the King’s Research Portal at https://kclpure.kcl.ac.uk/portal/ Strategies to increase -cell mass expansion Drynda, Robert Lech Awarding institution: King's College London The copyright of this thesis rests with the author and no quotation from it or information derived from it may be published without proper acknowledgement. END USER LICENCE AGREEMENT Unless another licence is stated on the immediately following page this work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International licence. https://creativecommons.org/licenses/by-nc-nd/4.0/ You are free to copy, distribute and transmit the work Under the following conditions: Attribution: You must attribute the work in the manner specified by the author (but not in any way that suggests that they endorse you or your use of the work). Non Commercial: You may not use this work for commercial purposes. No Derivative Works - You may not alter, transform, or build upon this work. Any of these conditions can be waived if you receive permission from the author. Your fair dealings and other rights are in no way affected by the above. Take down policy If you believe that this document breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 Strategies to increase β-cell mass expansion A thesis submitted by Robert Drynda For the degree of Doctor of Philosophy from King’s College London Diabetes Research Group Division of Diabetes & Nutritional Sciences Faculty of Life Sciences & Medicine King’s College London 2017 Table of contents Table of contents ................................................................................................. -

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

Receptor-Mediated Dimerization of JAK2 FERM Domains Is Required for JAK2 Activation Ryan D Ferrao, Heidi JA Wallweber, Patrick J Lupardus*
RESEARCH ARTICLE Receptor-mediated dimerization of JAK2 FERM domains is required for JAK2 activation Ryan D Ferrao, Heidi JA Wallweber, Patrick J Lupardus* Department of Structural Biology, Genentech, Inc., South San Francisco, United States Abstract Cytokines and interferons initiate intracellular signaling via receptor dimerization and activation of Janus kinases (JAKs). How JAKs structurally respond to changes in receptor conformation induced by ligand binding is not known. Here, we present two crystal structures of the human JAK2 FERM and SH2 domains bound to Leptin receptor (LEPR) and Erythropoietin receptor (EPOR), which identify a novel dimeric conformation for JAK2. This 2:2 JAK2/receptor dimer, observed in both structures, identifies a previously uncharacterized receptor interaction essential to dimer formation that is mediated by a membrane-proximal peptide motif called the ‘switch’ region. Mutation of the receptor switch region disrupts STAT phosphorylation but does not affect JAK2 binding, indicating that receptor-mediated formation of the JAK2 FERM dimer is required for kinase activation. These data uncover the structural and molecular basis for how a cytokine-bound active receptor dimer brings together two JAK2 molecules to stimulate JAK2 kinase activity. DOI: https://doi.org/10.7554/eLife.38089.001 Introduction Janus kinases (JAKs) are a family of multi-domain non-receptor tyrosine kinases responsible for pleio- tropic regulatory effects on growth, development, immune and hematopoietic signaling (Leonard and O’Shea, 1998). The JAK family consists of four conserved members, including JAK1, *For correspondence: [email protected] JAK2, JAK3, and TYK2, which are differentially activated in response to cytokine and interferon stim- ulation. JAKs are constitutively bound to the intracellular domains of their cognate cytokine signaling Competing interest: See receptors, and are activated after cytokine-mediated dimerization or rearrangement of these recep- page 18 tors establishes a productive receptor signaling complex (Haan et al., 2006). -

Bone-Specific Master Transcription Factor Runx2 Regulates Signaling and Metabolism Related Programs in Osteoprogenitors
ISSN 0233-7657. Biopolymers and Cell. 2010. Vol. 26. N 4 Bone-specific master transcription factor Runx2 regulates signaling and metabolism related programs in osteoprogenitors N. M. Teplyuk1, 2, V. I. Teplyuk2 1University of Massachusetts Medical School 55, Lake Ave North, 01655, Worcester, MA, USA 2Institute of Molecular Biology and Genetics of National Academy of Sciences of Ukraine 150, Zabolotnogo str., Kiev, Ukraine, 03680 [email protected] Aim. Runx2 (AML3) transcription factor is the key regulator of osteoblastic lineage progression and is indispensable for the formation of mineral bones. Runx2 expression increases during differentiation of osteoblasts to induce osteoblast-specific genes necessary for the production and deposition of bone mineral matrix. However, Runx2 is also expressed at a lower level in early osteoprogenitors, where its function is less understood. Here we study how Runx2 determines the early stages of osteoblastic commitment using the model system of Runx2 re-introduction in mouse calvaria cells with Runx2 null background. Method. Affymetrix analysis, Western blot analysis and quantitative real-time reverse transcriptase PCR (qRT-PCR) analysis were employed. Results. Gene expression profiling by Affymetrix microarrays revealed that along with the induction of extracellular matrix and bone mineral deposition related phenotypic markers, Runx2 regulates several cell programs related to signaling and metabolism in the early osteoprogenitors. Particularly, Runx2 regulates transcription of genes involved in G-protein coupled signaling network, FGF and BMP/TGF beta signaling pathways and in biogenesis and metabolism pathways of steroid hormones. Conclusion. The data indicate that the lineage specific program, regulated by the master regulatory transcription factor, includes the regulation of cellular signaling and metabolism which may allow the committed cell to react and behave differently in the same microenvironment. -
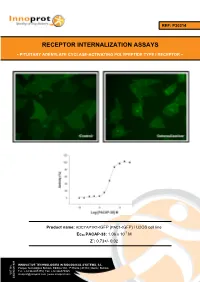
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Kinesin Family Member 18B Regulates the Proliferation and Invasion Of
Wu et al. Cell Death and Disease (2021) 12:302 https://doi.org/10.1038/s41419-021-03582-2 Cell Death & Disease ARTICLE Open Access Kinesin family member 18B regulates the proliferation and invasion of human prostate cancer cells Yu-Peng Wu 1,Zhi-BinKe 1, Wen-Cai Zheng 1, Ye-Hui Chen 1,Jun-MingZhu 1,FeiLin 1,Xiao-DongLi 1, Shao-Hao Chen 1,HaiCai 1, Qing-Shui Zheng 1, Yong Wei 1, Xue-Yi Xue 1 and Ning Xu 1 Abstract Expression of kinesin family member 18B (KIF18B), an ATPase with key roles in cell division, is deregulated in many cancers, but its involvement in prostate cancer (PCa) is unclear. Here, we investigated the expression and function of KIF18B in human PCa specimens and cell lines using bioinformatics analyses, immunohistochemical and immunofluorescence microscopy, and RT-qPCR and western blot analyses. KIF18B was overexpressed in PCa specimens compared with paracancerous tissues and was associated with poorer disease-free survival. In vitro, KIF18B knockdown in PCa cell lines promoted cell proliferation, migration, and invasion, and inhibited cell apoptosis, while KIF18B overexpression had the opposite effects. In a mouse xenograft model, KIF18B overexpression accelerated and promoted the growth of PCa tumors. Bioinformatics analysis of control and KIF18B-overexpressing PCa cells showed that genes involved in the PI3K–AKT–mTOR signaling pathway were significantly enriched among the differentially expressed genes. Consistent with this observation, we found that KIF18B overexpression activates the PI3K–AKT–mTOR signaling pathway in PCa cells both in vitro and in vivo. Collectively, our results suggest that KIF18B plays a crucial role – – 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; in PCa via activation of the PI3K AKT mTOR signaling pathway, and raise the possibility that KIF18B could have utility as a novel biomarker for PCa. -

Serum Levels of Spexin and Kisspeptin Negatively Correlate with Obesity and Insulin Resistance in Women
Physiol. Res. 67: 45-56, 2018 https://doi.org/10.33549/physiolres.933467 Serum Levels of Spexin and Kisspeptin Negatively Correlate With Obesity and Insulin Resistance in Women P. A. KOŁODZIEJSKI1, E. PRUSZYŃSKA-OSZMAŁEK1, E. KOREK4, M. SASSEK1, D. SZCZEPANKIEWICZ1, P. KACZMAREK1, L. NOGOWSKI1, P. MAĆKOWIAK1, K. W. NOWAK1, H. KRAUSS4, M. Z. STROWSKI2,3 1Department of Animal Physiology and Biochemistry, Poznan University of Life Sciences, Poznan, Poland, 2Department of Hepatology and Gastroenterology & The Interdisciplinary Centre of Metabolism: Endocrinology, Diabetes and Metabolism, Charité-University Medicine Berlin, Berlin, Germany, 3Department of Internal Medicine, Park-Klinik Weissensee, Berlin, Germany, 4Department of Physiology, Karol Marcinkowski University of Medical Science, Poznan, Poland Received August 18, 2016 Accepted June 19, 2017 On-line November 10, 2017 Summary Corresponding author Spexin (SPX) and kisspeptin (KISS) are novel peptides relevant in P. A. Kolodziejski, Department of Animal Physiology and the context of regulation of metabolism, food intake, puberty and Biochemistry, Poznan University of Life Sciences, Wolynska Street reproduction. Here, we studied changes of serum SPX and KISS 28, 60-637 Poznan, Poland. E-mail: [email protected] levels in female non-obese volunteers (BMI<25 kg/m2) and obese patients (BMI>35 kg/m2). Correlations between SPX or Introduction KISS with BMI, McAuley index, QUICKI, HOMA IR, serum levels of insulin, glucagon, leptin, adiponectin, orexin-A, obestatin, Kisspeptin (KISS) and spexin (SPX) are peptides ghrelin and GLP-1 were assessed. Obese patients had lower SPX involved in regulation of body weight, metabolism and and KISS levels as compared to non-obese volunteers (SPX: sexual functions. In 2014, Kim and coworkers showed that 4.48±0.19 ng/ml vs. -

Molecular Dissection of G-Protein Coupled Receptor Signaling and Oligomerization
MOLECULAR DISSECTION OF G-PROTEIN COUPLED RECEPTOR SIGNALING AND OLIGOMERIZATION BY MICHAEL RIZZO A Dissertation Submitted to the Graduate Faculty of WAKE FOREST UNIVERSITY GRADUATE SCHOOL OF ARTS AND SCIENCES in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY Biology December, 2019 Winston-Salem, North Carolina Approved By: Erik C. Johnson, Ph.D. Advisor Wayne E. Pratt, Ph.D. Chair Pat C. Lord, Ph.D. Gloria K. Muday, Ph.D. Ke Zhang, Ph.D. ACKNOWLEDGEMENTS I would first like to thank my advisor, Dr. Erik Johnson, for his support, expertise, and leadership during my time in his lab. Without him, the work herein would not be possible. I would also like to thank the members of my committee, Dr. Gloria Muday, Dr. Ke Zhang, Dr. Wayne Pratt, and Dr. Pat Lord, for their guidance and advice that helped improve the quality of the research presented here. I would also like to thank members of the Johnson lab, both past and present, for being valuable colleagues and friends. I would especially like to thank Dr. Jason Braco, Dr. Jon Fisher, Dr. Jake Saunders, and Becky Perry, all of whom spent a great deal of time offering me advice, proofreading grants and manuscripts, and overall supporting me through the ups and downs of the research process. Finally, I would like to thank my family, both for instilling in me a passion for knowledge and education, and for their continued support. In particular, I would like to thank my wife Emerald – I am forever indebted to you for your support throughout this process, and I will never forget the sacrifices you made to help me get to where I am today. -

0.5) in Stat3∆/∆ Compared with Stat3flox/Flox
Supplemental Table 2 Genes down-regulated (<0.5) in Stat3∆/∆ compared with Stat3flox/flox Probe ID Gene Symbol Gene Description Entrez gene ID 1460599_at Ermp1 endoplasmic reticulum metallopeptidase 1 226090 1460463_at H60c histocompatibility 60c 670558 1460431_at Gcnt1 glucosaminyl (N-acetyl) transferase 1, core 2 14537 1459979_x_at Zfp68 zinc finger protein 68 24135 1459747_at --- --- --- 1459608_at --- --- --- 1459168_at --- --- --- 1458718_at --- --- --- 1458618_at --- --- --- 1458466_at Ctsa cathepsin A 19025 1458345_s_at Colec11 collectin sub-family member 11 71693 1458046_at --- --- --- 1457769_at H60a histocompatibility 60a 15101 1457680_a_at Tmem69 transmembrane protein 69 230657 1457644_s_at Cxcl1 chemokine (C-X-C motif) ligand 1 14825 1457639_at Atp6v1h ATPase, H+ transporting, lysosomal V1 subunit H 108664 1457260_at 5730409E04Rik RIKEN cDNA 5730409E04Rik gene 230757 1457070_at --- --- --- 1456893_at --- --- --- 1456823_at Gm70 predicted gene 70 210762 1456671_at Tbrg3 transforming growth factor beta regulated gene 3 21378 1456211_at Nlrp10 NLR family, pyrin domain containing 10 244202 1455881_at Ier5l immediate early response 5-like 72500 1455576_at Rinl Ras and Rab interactor-like 320435 1455304_at Unc13c unc-13 homolog C (C. elegans) 208898 1455241_at BC037703 cDNA sequence BC037703 242125 1454866_s_at Clic6 chloride intracellular channel 6 209195 1453906_at Med13l mediator complex subunit 13-like 76199 1453522_at 6530401N04Rik RIKEN cDNA 6530401N04 gene 328092 1453354_at Gm11602 predicted gene 11602 100380944 1453234_at -

Galanin Stimulates Cortisol Secretion from Human Adrenocortical Cells
859-864 9/11/07 11:36 Page 859 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 20: 859-864, 2007 859 Galanin stimulates cortisol secretion from human adrenocortical cells through the activation of galanin receptor subtype 1 coupled to the adenylate cyclase-dependent signaling cascade ANNA S. BELLONI1, LUDWIK K. MALENDOWICZ2, MARCIN RUCINSKI2, DIEGO GUIDOLIN1 and GASTONE G. NUSSDORFER1 1Department of Human Anatomy and Physiology, School of Medicine, University of Padua, I-35121 Padua, Italy; 2Department of Histology and Embryology, Poznan School of Medicine, PL-60781 Poznan, Poland Received September 10, 2007; Accepted October 5, 2007 Abstract. Previous studies showed that galanin receptors are Introduction expressed in the rat adrenal, and galanin modulates gluco- corticoid secretion in this species. Hence, we investigated the Galanin is a regulatory peptide (30 amino acid residues in expression of the various galanin receptor subtypes (GAL-R1, humans) originally isolated from pig intestine (1) which is GAL-R2 and GAL-R3) in the human adrenocortical cells, and widely distributed in the central and peripheral nervous the possible involvement of galanin in the control of cortisol system, where it acts as a neurotransmitter/neuromodulator. secretion. Reverse transcription-polymerase chain reaction In the gut, galanin modulates insulin release and intestine detected the expression of GAL-R1 (but not GAL-R2 and contractility (2,3). Galanin acts through three distinct subtypes GAL-R3) in the inner zones of the human adrenal cortex. The of G protein-coupled receptors, referred to as GAL-R1, GAL-R2 galanin concentration dependently enhanced basal, but not and GAL-R3 (4). ACTH-stimulated secretion of cortisol from dispersed inner Evidence suggests that galanin is involved in the functional adrenocortical cells (maximal effective concentration, 10-8 M). -

Characterization of a High-Affinity Galanin Receptor in the Rat
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 4231-4235, May 1993 Neurobiology Characterization of a high-affinity galanin receptor in the rat anterior pituitary: Absence of biological effect and reduced membrane binding of the antagonist M15 differentiate it from the brain/gut receptor (galanin fragment/hemolytic plaque technique/prolactin) DAVID WYNICK*, DAVID M. SMITH*, MOHAMMAD GHATEI*, KAREN AKINSANYA*, RANJEV BHOGAL*, PAUL PURKISSt, PETER BYFIELDt, NOBORU YANAIHARAt, AND STEPHEN R. BLOOM* *Department of Medicine, Hammersmith Hospital, London W12 ONN, United Kingdom; tHaemostasis Research Group, Clinical Research Centre, Harrow, Middlesex, HAl 3UJ, United Kingdom; and tDepartment of Bio-organic Chemistry, University of Shizuoka, Shizuoka, Japan Communicated by L. L. Iversen, December 30, 1992 (received for review November 24, 1992) ABSTRACT Structure-activity studies demonstrate that anterior pituitary, where it has been shown to be estrogen galanin fragments 1-15 and 2-29 are fully active, whereas inducible (5). fragment 3-29 has been reported to be inactive, in a number Various studies have demonstrated effects of galanin on ofdifferent in vivo models. M15, a chimeric peptide comprising basal and stimulated release of prolactin (6, 7), growth galanin 1-13 and substance P 5-11, has recently been found to hormone (8-11), and luteinizing hormone (12, 13) either from be a potent galanin antagonist. Direct effects of galanin at the dispersed pituitary cells or at the hypothalamic level modu- level of the pituitary have been defined, yet, paradoxically, a lating dopamine, somatostatin (SRIF; somatotropin release- number of studies have been unable to demonstrate galanin inhibiting factor), and gonadotropin-releasing hormone binding to an anterior pituitary receptor.