Gene Standard Deviation MTOR 0.12553731 PRPF38A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

PRODUCT SPECIFICATION Anti-EIF2B4 Product
Anti-EIF2B4 Product Datasheet Polyclonal Antibody PRODUCT SPECIFICATION Product Name Anti-EIF2B4 Product Number HPA039993 Gene Description eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa Clonality Polyclonal Isotype IgG Host Rabbit Antigen Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: VGREMTKEEKLQLRKEKKQQKKKRKEEKGAEPETGSAVSAAQCQVGPTRE LPESGIQLGTPREKVPAGRSKAELRAER Purification Method Affinity purified using the PrEST antigen as affinity ligand Verified Species Human Reactivity Recommended ICC-IF (Immunofluorescence) Applications - Fixation/Permeabilization: PFA/Triton X-100 - Working concentration: 0.25-2 µg/ml Characterization Data Available at atlasantibodies.com/products/HPA039993 Buffer 40% glycerol and PBS (pH 7.2). 0.02% sodium azide is added as preservative. Concentration Lot dependent Storage Store at +4°C for short term storage. Long time storage is recommended at -20°C. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. For protocols, additional product information, such as images and references, see atlasantibodies.com. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -

Supplementary Materials: Evaluation of Cytotoxicity and Α-Glucosidase Inhibitory Activity of Amide and Polyamino-Derivatives of Lupane Triterpenoids
Supplementary Materials: Evaluation of cytotoxicity and α-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids Oxana B. Kazakova1*, Gul'nara V. Giniyatullina1, Akhat G. Mustafin1, Denis A. Babkov2, Elena V. Sokolova2, Alexander A. Spasov2* 1Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences, 71, pr. Oktyabrya, 450054 Ufa, Russian Federation 2Scientific Center for Innovative Drugs, Volgograd State Medical University, Novorossiyskaya st. 39, Volgograd 400087, Russian Federation Correspondence Prof. Dr. Oxana B. Kazakova Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences 71 Prospeсt Oktyabrya Ufa, 450054 Russian Federation E-mail: [email protected] Prof. Dr. Alexander A. Spasov Scientific Center for Innovative Drugs of the Volgograd State Medical University 39 Novorossiyskaya st. Volgograd, 400087 Russian Federation E-mail: [email protected] Figure S1. 1H and 13C of compound 2. H NH N H O H O H 2 2 Figure S2. 1H and 13C of compound 4. NH2 O H O H CH3 O O H H3C O H 4 3 Figure S3. Anticancer screening data of compound 2 at single dose assay 4 Figure S4. Anticancer screening data of compound 7 at single dose assay 5 Figure S5. Anticancer screening data of compound 8 at single dose assay 6 Figure S6. Anticancer screening data of compound 9 at single dose assay 7 Figure S7. Anticancer screening data of compound 12 at single dose assay 8 Figure S8. Anticancer screening data of compound 13 at single dose assay 9 Figure S9. Anticancer screening data of compound 14 at single dose assay 10 Figure S10. -

Neprilysin Is Required for Angiotensin-(1-7)
Page 1 of 39 Diabetes NEPRILYSIN IS REQUIRED FOR ANGIOTENSIN-(1-7)’S ABILITY TO ENHANCE INSULIN SECRETION VIA ITS PROTEOLYTIC ACTIVITY TO GENERATE ANGIOTENSIN-(1-2) Gurkirat S. Brara, Breanne M. Barrowa, Matthew Watsonb, Ryan Griesbachc, Edwina Chounga, Andrew Welchc, Bela Ruzsicskad, Daniel P. Raleighb, Sakeneh Zraikaa,c aVeterans Affairs Puget Sound Health Care System, Seattle, WA 98108, United States bDepartment of Chemistry, Stony Brook University, Stony Brook, NY 11794, United States cDivision of Metabolism, Endocrinology and Nutrition, Department of Medicine, University of Washington, Seattle, WA 98195, United States dInstitute for Chemical Biology and Drug Discovery, Stony Brook University, Stony Brook, NY 11794, United States Short Title: Angiotensin-(1-7) and insulin secretion Word count: 3997; Figure count: 8 main (plus 3 Online Suppl.); Table count: 1 Online Suppl. Correspondence to: Sakeneh Zraika, PhD 1660 South Columbian Way (151) Seattle, WA, United States Tel: 206-768-5391 / Fax: 206-764-2164 Email: [email protected] 1 Diabetes Publish Ahead of Print, published online May 30, 2017 Diabetes Page 2 of 39 ABSTRACT Recent work has renewed interest in therapies targeting the renin-angiotensin system (RAS) to improve β-cell function in type 2 diabetes. Studies show that generation of angiotensin-(1-7) by angiotensin converting enzyme 2 (ACE2) and its binding to the Mas receptor (MasR) improves glucose homeostasis, partly by enhancing glucose-stimulated insulin secretion (GSIS). Thus, islet ACE2 upregulation is viewed as a desirable therapeutic goal. Here, we show that although endogenous islet ACE2 expression is sparse, its inhibition abrogates angiotensin-(1-7)-mediated GSIS. However, a more widely expressed islet peptidase, neprilysin, degrades angiotensin-(1-7) into several peptides. -

Familial Juvenile Polyposis Syndrome with a De Novo Germline Missense Variant in BMPR1A Gene: a Case Report Qing Liu, Mengling Liu, Tianshu Liu and Yiyi Yu*
Liu et al. BMC Medical Genetics (2020) 21:196 https://doi.org/10.1186/s12881-020-01135-6 CASE REPORT Open Access Familial juvenile polyposis syndrome with a de novo germline missense variant in BMPR1A gene: a case report Qing Liu, Mengling Liu, Tianshu Liu and Yiyi Yu* Abstract Background: Juvenile polyposis syndrome (JPS) is a rare autosomal dominant hereditary disorder characterized by the development of multiple distinct juvenile polyps in the gastrointestinal tract with an increased risk of colorectal cancer. Germline mutations in two genes, SMAD4 and BMPR1A, have been identified to cause JPS. Case presentation: Here, we report a germline heterozygous missense variant (c.299G > A) in exon 3 BMPR1A gene in a family with juvenile polyposis. This variant was absent from the population database, and concluded as de novo compared with the parental sequencing. Further sequencing of the proband’s children confirmed the segregation of this variant with the disease, while the variant was also predicted to have damaging effect based on online prediction tools. Therefore, this variant was classified as likely pathogenic according to the American College of Medical Genetics and Genomics (ACMG) guidelines. Conclusions: Germline genetic testing revealed a de novo germline missense variant in BMPR1A gene in a family with juvenile polyposis. Identification of the pathogenic variant facilitates the cancer risk management of at-risk family members, and endoscopic surveillance is recommended for mutation carriers. Keywords: Juvenile polyposis syndrome, BMPR1A gene, De novo germline variant, Missense variant Background two genes, SMAD4 and BMPR1A, have been identi- Juvenile polyposis syndrome (JPS) is a rare autosomal fied to cause JPS [5]. -

The Prevalence of MADH4 and BMPR1A Mutations in Juvenile Polyposis and Absence of BMPR2, BMPR1B, and ACVR1 Mutations
484 ORIGINAL ARTICLE J Med Genet: first published as 10.1136/jmg.2004.018598 on 2 July 2004. Downloaded from The prevalence of MADH4 and BMPR1A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations J R Howe, M G Sayed, A F Ahmed, J Ringold, J Larsen-Haidle, A Merg, F A Mitros, C A Vaccaro, G M Petersen, F M Giardiello, S T Tinley, L A Aaltonen, H T Lynch ............................................................................................................................... J Med Genet 2004;41:484–491. doi: 10.1136/jmg.2004.018598 Background: Juvenile polyposis (JP) is an autosomal dominant syndrome predisposing to colorectal and gastric cancer. We have identified mutations in two genes causing JP, MADH4 and bone morphogenetic protein receptor 1A (BMPR1A): both are involved in bone morphogenetic protein (BMP) mediated signalling and are members of the TGF-b superfamily. This study determined the prevalence of mutations See end of article for in MADH4 and BMPR1A, as well as three other BMP/activin pathway candidate genes in a large number authors’ affiliations ....................... of JP patients. Methods: DNA was extracted from the blood of JP patients and used for PCR amplification of each exon of Correspondence to: these five genes, using primers flanking each intron–exon boundary. Mutations were determined by Dr J R Howe, Department of Surgery, 4644 JCP, comparison to wild type sequences using sequence analysis software. A total of 77 JP cases were University of Iowa College sequenced for mutations in the MADH4, BMPR1A, BMPR1B, BMPR2, and/or ACVR1 (activin A receptor) of Medicine, 200 Hawkins genes. The latter three genes were analysed when MADH4 and BMPR1A sequencing found no mutations. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Download Download
Supplementary Figure S1. Results of flow cytometry analysis, performed to estimate CD34 positivity, after immunomagnetic separation in two different experiments. As monoclonal antibody for labeling the sample, the fluorescein isothiocyanate (FITC)- conjugated mouse anti-human CD34 MoAb (Mylteni) was used. Briefly, cell samples were incubated in the presence of the indicated MoAbs, at the proper dilution, in PBS containing 5% FCS and 1% Fc receptor (FcR) blocking reagent (Miltenyi) for 30 min at 4 C. Cells were then washed twice, resuspended with PBS and analyzed by a Coulter Epics XL (Coulter Electronics Inc., Hialeah, FL, USA) flow cytometer. only use Non-commercial 1 Supplementary Table S1. Complete list of the datasets used in this study and their sources. GEO Total samples Geo selected GEO accession of used Platform Reference series in series samples samples GSM142565 GSM142566 GSM142567 GSM142568 GSE6146 HG-U133A 14 8 - GSM142569 GSM142571 GSM142572 GSM142574 GSM51391 GSM51392 GSE2666 HG-U133A 36 4 1 GSM51393 GSM51394 only GSM321583 GSE12803 HG-U133A 20 3 GSM321584 2 GSM321585 use Promyelocytes_1 Promyelocytes_2 Promyelocytes_3 Promyelocytes_4 HG-U133A 8 8 3 GSE64282 Promyelocytes_5 Promyelocytes_6 Promyelocytes_7 Promyelocytes_8 Non-commercial 2 Supplementary Table S2. Chromosomal regions up-regulated in CD34+ samples as identified by the LAP procedure with the two-class statistics coded in the PREDA R package and an FDR threshold of 0.5. Functional enrichment analysis has been performed using DAVID (http://david.abcc.ncifcrf.gov/) -

Cell-Autonomous FLT3L Shedding Via ADAM10 Mediates Conventional Dendritic Cell Development in Mouse Spleen
Cell-autonomous FLT3L shedding via ADAM10 mediates conventional dendritic cell development in mouse spleen Kohei Fujitaa,b,1, Svetoslav Chakarovc,1, Tetsuro Kobayashid, Keiko Sakamotod, Benjamin Voisind, Kaibo Duanc, Taneaki Nakagawaa, Keisuke Horiuchie, Masayuki Amagaib, Florent Ginhouxc, and Keisuke Nagaod,2 aDepartment of Dentistry and Oral Surgery, Keio University School of Medicine, Tokyo 160-8582, Japan; bDepartment of Dermatology, Keio University School of Medicine, Tokyo 160-8582, Japan; cSingapore Immunology Network, Agency for Science, Technology and Research, Biopolis, 138648 Singapore; dDermatology Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892; and eDepartment of Orthopedic Surgery, National Defense Medical College, Tokorozawa 359-8513, Japan Edited by Kenneth M. Murphy, Washington University School of Medicine, St. Louis, MO, and approved June 10, 2019 (received for review November 4, 2018) Conventional dendritic cells (cDCs) derive from bone marrow (BM) intocDC1sorcDC2stakesplaceintheBM(3),andthese precursors that undergo cascades of developmental programs to pre-cDC1s and pre-cDC2s ultimately differentiate into cDC1s terminally differentiate in peripheral tissues. Pre-cDC1s and pre- and cDC2s after migrating to nonlymphoid and lymphoid tissues. + + cDC2s commit in the BM to each differentiate into CD8α /CD103 cDCs are short-lived, and their homeostatic maintenance relies + cDC1s and CD11b cDC2s, respectively. Although both cDCs rely on on constant replenishment from the BM precursors (5). The cy- the cytokine FLT3L during development, mechanisms that ensure tokine Fms-related tyrosine kinase 3 ligand (FLT3L) (12), by cDC accessibility to FLT3L have yet to be elucidated. Here, we gen- signaling through its receptor FLT3 expressed on DC precursors, erated mice that lacked a disintegrin and metalloproteinase (ADAM) is essential during the development of DCs (7, 13). -

Tumor Promoting Effect of BMP Signaling in Endometrial Cancer
International Journal of Molecular Sciences Article Tumor Promoting Effect of BMP Signaling in Endometrial Cancer Tomohiko Fukuda 1,* , Risa Fukuda 1, Kohei Miyazono 1,2,† and Carl-Henrik Heldin 1,*,† 1 Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Box 582, Uppsala University, SE-751 23 Uppsala, Sweden; [email protected] (R.F.); [email protected] (K.M.) 2 Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Tokyo 113-0033, Japan * Correspondence: [email protected] (T.F.); [email protected] (C.-H.H.); Tel.: +46-18-4714738 (T.F.); +46-18-4714738 (C.-H.H.) † These authors contributed equally to this work. Abstract: The effects of bone morphogenetic proteins (BMPs), members of the transforming growth factor-β (TGF-β) family, in endometrial cancer (EC) have yet to be determined. In this study, we analyzed the TCGA and MSK-IMPACT datasets and investigated the effects of BMP2 and of TWSG1, a BMP antagonist, on Ishikawa EC cells. Frequent ACVR1 mutations and high mRNA expressions of BMP ligands and receptors were observed in EC patients of the TCGA and MSK-IMPACT datasets. Ishikawa cells secreted higher amounts of BMP2 compared with ovarian cancer cell lines. Exogenous BMP2 stimulation enhanced EC cell sphere formation via c-KIT induction. BMP2 also induced EMT of EC cells, and promoted migration by induction of SLUG. The BMP receptor kinase inhibitor LDN193189 augmented the growth inhibitory effects of carboplatin. Analyses of mRNAs of several BMP antagonists revealed that TWSG1 mRNA was abundantly expressed in Ishikawa cells. -

Mirna Let-7 from TPO(+) Extracellular Vesicles Is a Potential Marker for a Differential Diagnosis of Follicular Thyroid Nodules
cells Article MiRNA let-7 from TPO(+) Extracellular Vesicles is a Potential Marker for a Differential Diagnosis of Follicular Thyroid Nodules Lidia Zabegina 1,2,3, Inga Nazarova 1,2, Margarita Knyazeva 1,2,3, Nadezhda Nikiforova 1,2, Maria Slyusarenko 1,2, Sergey Titov 4 , Dmitry Vasilyev 1, Ilya Sleptsov 5 and Anastasia Malek 1,2,* 1 Subcellular Technology Lab., N.N. Petrov National Medical Research Center of Oncology, 195251 St. Petersburg, Russia; [email protected] (L.Z.); [email protected] (I.N.); [email protected] (M.K.); [email protected] (N.N.); [email protected] (M.S.); [email protected] (D.V.) 2 Oncosystem Ltd., 121205 Moscow, Russia 3 Institute of Biomedical Systems and Biotechnologies, Peter the Great St. Petersburg Polytechnic University, 195251 St. Petersburg, Russia 4 PCR Laboratory; AO Vector-Best, 630117 Novosibirsk, Russia; [email protected] 5 Department of endocrine surgery, Clinic of High Medical Technologies, St. Petersburg State University N.I. Pirogov, 190103 St. Petersburg, Russia; [email protected] * Correspondence: [email protected]; Tel.: +7-960-250-46-80 Received: 19 July 2020; Accepted: 15 August 2020; Published: 18 August 2020 Abstract: Background: The current approaches to distinguish follicular adenomas (FA) and follicular thyroid cancer (FTC) at the pre-operative stage have low predictive value. Liquid biopsy-based analysis of circulating extracellular vesicles (EVs) presents a promising diagnostic method. However, the extreme heterogeneity of plasma EV population hampers the development of new diagnostic tests. We hypothesize that the isolation of EVs with thyroid-specific surface molecules followed by miRNA analysis, may have improved diagnostic potency. -
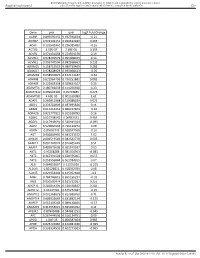
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al.