G Protein-Coupled Receptors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mglu2 Receptor Agonism, but Not Positive Allosteric Modulation, Elicits Rapid Tolerance Towards Their Primary Efficacy on Sleep Measures in Rats
RESEARCH ARTICLE mGlu2 Receptor Agonism, but Not Positive Allosteric Modulation, Elicits Rapid Tolerance towards Their Primary Efficacy on Sleep Measures in Rats Abdallah Ahnaou1*, Hilde Lavreysen1, Gary Tresadern2, Jose M. Cid2, Wilhelmus H. Drinkenburg1 1 Dept. of Neuroscience, Janssen Research & Development, A Division of Janssen Pharmaceutica N.V., Turnhoutseweg 30, B-2340, Beerse, Belgium, 2 Neuroscience Medicinal Chemistry, Janssen Research & Development, Janssen-Cilag S.A., Jarama 75, Polígono Industrial, 45007, Toledo, Spain * [email protected] Abstract OPEN ACCESS G-protein-coupled receptor (GPCR) agonists are known to induce both cellular adaptations Citation: Ahnaou A, Lavreysen H, Tresadern G, Cid resulting in tolerance to therapeutic effects and withdrawal symptoms upon treatment dis- JM, Drinkenburg WH (2015) mGlu2 Receptor continuation. Glutamate neurotransmission is an integral part of sleep-wake mechanisms, Agonism, but Not Positive Allosteric Modulation, Elicits Rapid Tolerance towards Their Primary which processes have translational relevance for central activity and target engagement. Efficacy on Sleep Measures in Rats. PLoS ONE 10 Here, we investigated the efficacy and tolerance potential of the metabotropic glutamate (12): e0144017. doi:10.1371/journal.pone.0144017 receptors (mGluR2/3) agonist LY354740 versus mGluR2 positive allosteric modulator Editor: James Porter, University of North Dakota, (PAM) JNJ-42153605 on sleep-wake organisation in rats. In vitro, the selectivity and UNITED STATES potency of JNJ-42153605 were characterized. In vivo, effects on sleep measures were Received: July 12, 2015 investigated in rats after once daily oral repeated treatment for 7 days, withdrawal and con- Accepted: November 12, 2015 secutive re-administration of LY354740 (1–10 mg/kg) and JNJ-42153605 (3–30 mg/kg). -

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

Nitrate Prodrugs Able to Release Nitric Oxide in a Controlled and Selective
Europäisches Patentamt *EP001336602A1* (19) European Patent Office Office européen des brevets (11) EP 1 336 602 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.7: C07C 205/00, A61K 31/00 20.08.2003 Bulletin 2003/34 (21) Application number: 02425075.5 (22) Date of filing: 13.02.2002 (84) Designated Contracting States: (71) Applicant: Scaramuzzino, Giovanni AT BE CH CY DE DK ES FI FR GB GR IE IT LI LU 20052 Monza (Milano) (IT) MC NL PT SE TR Designated Extension States: (72) Inventor: Scaramuzzino, Giovanni AL LT LV MK RO SI 20052 Monza (Milano) (IT) (54) Nitrate prodrugs able to release nitric oxide in a controlled and selective way and their use for prevention and treatment of inflammatory, ischemic and proliferative diseases (57) New pharmaceutical compounds of general effects and for this reason they are useful for the prep- formula (I): F-(X)q where q is an integer from 1 to 5, pref- aration of medicines for prevention and treatment of in- erably 1; -F is chosen among drugs described in the text, flammatory, ischemic, degenerative and proliferative -X is chosen among 4 groups -M, -T, -V and -Y as de- diseases of musculoskeletal, tegumental, respiratory, scribed in the text. gastrointestinal, genito-urinary and central nervous sys- The compounds of general formula (I) are nitrate tems. prodrugs which can release nitric oxide in vivo in a con- trolled and selective way and without hypotensive side EP 1 336 602 A1 Printed by Jouve, 75001 PARIS (FR) EP 1 336 602 A1 Description [0001] The present invention relates to new nitrate prodrugs which can release nitric oxide in vivo in a controlled and selective way and without the side effects typical of nitrate vasodilators drugs. -
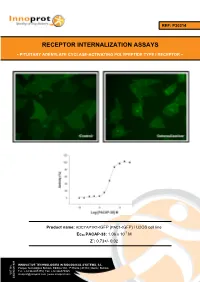
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Metabotropic Glutamate Receptors
mGluR Metabotropic glutamate receptors mGluR (metabotropic glutamate receptor) is a type of glutamate receptor that are active through an indirect metabotropic process. They are members of thegroup C family of G-protein-coupled receptors, or GPCRs. Like all glutamate receptors, mGluRs bind with glutamate, an amino acid that functions as an excitatoryneurotransmitter. The mGluRs perform a variety of functions in the central and peripheral nervous systems: mGluRs are involved in learning, memory, anxiety, and the perception of pain. mGluRs are found in pre- and postsynaptic neurons in synapses of the hippocampus, cerebellum, and the cerebral cortex, as well as other parts of the brain and in peripheral tissues. Eight different types of mGluRs, labeled mGluR1 to mGluR8, are divided into groups I, II, and III. Receptor types are grouped based on receptor structure and physiological activity. www.MedChemExpress.com 1 mGluR Agonists, Antagonists, Inhibitors, Modulators & Activators (-)-Camphoric acid (1R,2S)-VU0155041 Cat. No.: HY-122808 Cat. No.: HY-14417A (-)-Camphoric acid is the less active enantiomer (1R,2S)-VU0155041, Cis regioisomer of VU0155041, is of Camphoric acid. Camphoric acid stimulates a partial mGluR4 agonist with an EC50 of 2.35 osteoblast differentiation and induces μM. glutamate receptor expression. Camphoric acid also significantly induced the activation of NF-κB and AP-1. Purity: ≥98.0% Purity: ≥98.0% Clinical Data: No Development Reported Clinical Data: No Development Reported Size: 10 mM × 1 mL, 100 mg Size: 10 mM × 1 mL, 5 mg, 10 mg, 25 mg (2R,4R)-APDC (R)-ADX-47273 Cat. No.: HY-102091 Cat. No.: HY-13058B (2R,4R)-APDC is a selective group II metabotropic (R)-ADX-47273 is a potent mGluR5 positive glutamate receptors (mGluRs) agonist. -

The G Protein-Coupled Glutamate Receptors As Novel Molecular Targets in Schizophrenia Treatment— a Narrative Review
Journal of Clinical Medicine Review The G Protein-Coupled Glutamate Receptors as Novel Molecular Targets in Schizophrenia Treatment— A Narrative Review Waldemar Kryszkowski 1 and Tomasz Boczek 2,* 1 General Psychiatric Ward, Babinski Memorial Hospital in Lodz, 91229 Lodz, Poland; [email protected] 2 Department of Molecular Neurochemistry, Medical University of Lodz, 92215 Lodz, Poland * Correspondence: [email protected] Abstract: Schizophrenia is a severe neuropsychiatric disease with an unknown etiology. The research into the neurobiology of this disease led to several models aimed at explaining the link between perturbations in brain function and the manifestation of psychotic symptoms. The glutamatergic hypothesis postulates that disrupted glutamate neurotransmission may mediate cognitive and psychosocial impairments by affecting the connections between the cortex and the thalamus. In this regard, the greatest attention has been given to ionotropic NMDA receptor hypofunction. However, converging data indicates metabotropic glutamate receptors as crucial for cognitive and psychomotor function. The distribution of these receptors in the brain regions related to schizophrenia and their regulatory role in glutamate release make them promising molecular targets for novel antipsychotics. This article reviews the progress in the research on the role of metabotropic glutamate receptors in schizophrenia etiopathology. Citation: Kryszkowski, W.; Boczek, T. The G Protein-Coupled Glutamate Keywords: schizophrenia; metabotropic glutamate receptors; positive allosteric modulators; negative Receptors as Novel Molecular Targets allosteric modulators; drug development; animal models of schizophrenia; clinical trials in Schizophrenia Treatment—A Narrative Review. J. Clin. Med. 2021, 10, 1475. https://doi.org/10.3390/ jcm10071475 1. Introduction Academic Editors: Andreas Reif, Schizophrenia is a common debilitating disease affecting about 0.3–1% of the human Blazej Misiak and Jerzy Samochowiec population worldwide [1]. -

The 'C3ar Antagonist' SB290157 Is a Partial C5ar2 Agonist
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.01.232090; this version posted August 3, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The ‘C3aR antagonist’ SB290157 is a partial C5aR2 agonist Xaria X. Li1, Vinod Kumar1, John D. Lee1, Trent M. Woodruff1* 1School of Biomedical Sciences, The University of Queensland, St Lucia, 4072 Australia. * Correspondence: Prof. Trent M. Woodruff School of Biomedical Sciences, The University of Queensland, St Lucia, 4072 Australia. Ph: +61 7 3365 2924; Fax: +61 7 3365 1766; E-mail: [email protected] Keywords: Complement C3a, C3aR, SB290157, C5aR1, C5aR2 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.01.232090; this version posted August 3, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abbreviations used in this article: BRET, bioluminescence resonance energy transfer; BSA, bovine serum albumin; C3aR, C3a receptor C5aR1, C5a receptor 1; CHO-C3aR, Chinese hamster ovary cells stably expressing C3aR; CHO-C5aR1, Chinese hamster ovary cells stably expressing C5aR1; DMEM, Dulbecco's Modified Eagle's Medium; ERK1/2, extracellular signal-regulated kinase 1/2; FBS, foetal bovine serum; HEK293, human embryonic kidney 293 cells; HMDM, human monocyte-derived macrophage; i.p., intraperitoneal; i.v., intravenous; rhC5a, recombinant human C5a; RT, room temperature; S.E.M. -

The Impact of the Nonpeptide Corticotropin-Releasing Hormone Antagonist Antalarmin on Behavioral and Endocrine Responses to Stress*
0013-7227/99/$03.00/0 Vol. 140, No. 1 Endocrinology Printed in U.S.A. Copyright © 1999 by The Endocrine Society The Impact of the Nonpeptide Corticotropin-Releasing Hormone Antagonist Antalarmin on Behavioral and Endocrine Responses to Stress* TERRENCE DEAK, KIEN T. NGUYEN, ANDREA L. EHRLICH, LINDA R. WATKINS, ROBERT L. SPENCER, STEVEN F. MAIER, JULIO LICINIO, MA-LI WONG, GEORGE P. CHROUSOS, ELIZABETH WEBSTER, AND PHILIP W. GOLD Department of Psychology (T.D., K.T.N., A.L.E., L.R.W., R.L.S., S.F.M.), University of Colorado, Boulder, Colorado 80309-0345; Clinical Neuroendocrinology Branch (J.L., M.-L.W., P.W.G.), National Institute of Mental Health, National Institutes of Health, Bethesda, Maryland 20892-1284; and Developmental Neuroendocrinology Branch (G.P.C., E.W.), National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland 20892-1284 ABSTRACT Furthermore, because rats previously exposed to inescapable shock The nonpeptide CRH antagonist antalarmin has been shown to (IS; 100 shocks, 1.6 mA, 5 sec each), demonstrate enhanced fear block both behavioral and endocrine responses to CRH. However, it’s conditioning, we investigated whether this effect would be blocked by potential activity in blunting behavioral and endocrine sequelae of antalarmin. Antalarmin (20 mg/kgz2 ml ip) impaired both the induc- stressor exposure has not been assessed. Because antagonism of cen- tion and expression of conditioned fear. In addition, antalarmin tral CRH by a-helical CRH attenuates conditioned fear responses, we blocked the enhancement of fear conditioning produced by prior ex- sought to test antalarmin in this regard. -

Adverse Effects of Stress on Drug Addiction
Making a bad thing worse: adverse effects of stress on drug addiction Jessica N. Cleck, Julie A. Blendy J Clin Invest. 2008;118(2):454-461. https://doi.org/10.1172/JCI33946. Review Series Sustained exposure to various psychological stressors can exacerbate neuropsychiatric disorders, including drug addiction. Addiction is a chronic brain disease in which individuals cannot control their need for drugs, despite negative health and social consequences. The brains of addicted individuals are altered and respond very differently to stress than those of individuals who are not addicted. In this Review, we highlight some of the common effects of stress and drugs of abuse throughout the addiction cycle. We also discuss both animal and human studies that suggest treating the stress- related aspects of drug addiction is likely to be an important contributing factor to a long-lasting recovery from this disorder. Find the latest version: https://jci.me/33946/pdf Review series Making a bad thing worse: adverse effects of stress on drug addiction Jessica N. Cleck and Julie A. Blendy Department of Pharmacology, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA. Sustained exposure to various psychological stressors can exacerbate neuropsychiatric disorders, including drug addiction. Addiction is a chronic brain disease in which individuals cannot control their need for drugs, despite negative health and social consequences. The brains of addicted individuals are altered and respond very differently to stress than those of individuals who are not addicted. In this Review, we highlight some of the common effects of stress and drugs of abuse throughout the addiction cycle. -

BD Biosciences New RUO Reagents - November 2020
BD Biosciences New RUO reagents - November 2020 Reactivity Description Format Clone Size Cat. number Hu CD133 FITC W6B3C1 100µg 567029 Hu CD133 FITC W6B3C1 25µg 567033 Hu CD39 PE A1/CD39 100Tst 567156 Hu CD39 PE A1/CD39 25Tst 567157 Hu KIR2DL1/S1/S3/S5 PE HP-MA4 100Tst 567158 Hu KIR2DL1/S1/S3/S5 PE HP-MA4 25Tst 567159 Hu IL-22 Alexa Fluor® 647 MH22B2 100µg 567160 Hu IL-22 Alexa Fluor® 647 MH22B2 25µg 567161 Hu CD99 R718 TU12 50µg 751651 Hu CD161 R718 DX12 50µg 751652 Hu CD116 R718 HGMCSFR-M1 50µg 751653 Hu HLA-G R718 87G 50µg 751670 Hu CD27 R718 O323 50µg 751686 Hu CD80 (B7-1) R718 2D10.4 50µg 751737 Hu Integrin αvβ5 R718 ALULA 50µg 751738 Hu CD266 (Tweak-R) R718 ITEM-4 50µg 751739 Hu ErbB3 (HER-3) R718 SGP1 50µg 751799 Hu TCR Vβ5.1 R718 LC4 50µg 751816 Hu CD123 (IL-3Ra) R718 6H6 50µg 751844 Hu CD1a R718 SK9 50µg 751847 Hu CD20 R718 L27 50µg 751849 Hu Disial GD2 R718 14.G2A 50µg 751851 Reactivity Description Format Clone Size Cat. number Hu CD71 R718 L01.1 50µg 751853 Hu CD278 (ICOS) R718 DX29 50µg 751854 Hu B7-H4 R718 MIH43 50µg 751857 Hu CD53 R718 HI29 50µg 751858 Hu CD197 (CCR7) R718 2-L1-A 50µg 751859 Hu CD197 (CCR7) R718 3D12 50µg 751861 Hu CD31 R718 L133.1 50µg 751863 Hu EGF Receptor R718 EMAB-134 50µg 751864 Hu CD8b R718 2ST8.5H7 50µg 751867 Hu CD31 R718 MBC 78.2 50µg 751869 Hu CD162 R718 KPL-1 50µg 751873 Hu CD24 R718 ML5 50µg 751874 Hu CD159C (NKG2C) R718 134591 50µg 751876 Hu CD169 (Siglec-1) R718 7-239 50µg 751877 Hu CD16b R718 CLB-GRAN11.5 50µg 751880 Hu IgM R718 UCH-B1 50µg 751881 Hu CD275 R718 2D3/B7-H2 50µg 751883 Hu CD307e -

Classification Decisions Taken by the Harmonized System Committee from the 47Th to 60Th Sessions (2011
CLASSIFICATION DECISIONS TAKEN BY THE HARMONIZED SYSTEM COMMITTEE FROM THE 47TH TO 60TH SESSIONS (2011 - 2018) WORLD CUSTOMS ORGANIZATION Rue du Marché 30 B-1210 Brussels Belgium November 2011 Copyright © 2011 World Customs Organization. All rights reserved. Requests and inquiries concerning translation, reproduction and adaptation rights should be addressed to [email protected]. D/2011/0448/25 The following list contains the classification decisions (other than those subject to a reservation) taken by the Harmonized System Committee ( 47th Session – March 2011) on specific products, together with their related Harmonized System code numbers and, in certain cases, the classification rationale. Advice Parties seeking to import or export merchandise covered by a decision are advised to verify the implementation of the decision by the importing or exporting country, as the case may be. HS codes Classification No Product description Classification considered rationale 1. Preparation, in the form of a powder, consisting of 92 % sugar, 6 % 2106.90 GRIs 1 and 6 black currant powder, anticaking agent, citric acid and black currant flavouring, put up for retail sale in 32-gram sachets, intended to be consumed as a beverage after mixing with hot water. 2. Vanutide cridificar (INN List 100). 3002.20 3. Certain INN products. Chapters 28, 29 (See “INN List 101” at the end of this publication.) and 30 4. Certain INN products. Chapters 13, 29 (See “INN List 102” at the end of this publication.) and 30 5. Certain INN products. Chapters 28, 29, (See “INN List 103” at the end of this publication.) 30, 35 and 39 6. Re-classification of INN products. -

Conformational Dynamics Between Transmembrane Domains And
RESEARCH ARTICLE Conformational dynamics between transmembrane domains and allosteric modulation of a metabotropic glutamate receptor Vanessa A Gutzeit1, Jordana Thibado2, Daniel Starer Stor2, Zhou Zhou3, Scott C Blanchard2,3,4, Olaf S Andersen2,3, Joshua Levitz2,4,5* 1Neuroscience Graduate Program, Weill Cornell Graduate School of Medical Sciences, New York, United States; 2Physiology, Biophysics and Systems Biology Graduate Program, Weill Cornell Graduate School of Medical Sciences, New York, United States; 3Department of Physiology and Biophysics, Weill Cornell Medicine, New York, United States; 4Tri-Institutional PhD Program in Chemical Biology, New York, United States; 5Department of Biochemistry, Weill Cornell Medicine, New York, United States Abstract Metabotropic glutamate receptors (mGluRs) are class C, synaptic G-protein-coupled receptors (GPCRs) that contain large extracellular ligand binding domains (LBDs) and form constitutive dimers. Despite the existence of a detailed picture of inter-LBD conformational dynamics and structural snapshots of both isolated domains and full-length receptors, it remains unclear how mGluR activation proceeds at the level of the transmembrane domains (TMDs) and how TMD-targeting allosteric drugs exert their effects. Here, we use time-resolved functional and conformational assays to dissect the mechanisms by which allosteric drugs activate and modulate mGluR2. Single-molecule subunit counting and inter-TMD fluorescence resonance energy transfer *For correspondence: measurements in living cells