GPCR Expression Profiles Were Determined Using
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Evidence for Gliadin Antibodies As Causative Agents in Schizophrenia
1 Evidence for gliadin antibodies as causative agents in schizophrenia. C.J.Carter PolygenicPathways, 20 Upper Maze Hill, Saint-Leonard’s on Sea, East Sussex, TN37 0LG [email protected] Tel: 0044 (0)1424 422201 I have no fax Abstract Antibodies to gliadin, a component of gluten, have frequently been reported in schizophrenia patients, and in some cases remission has been noted following the instigation of a gluten free diet. Gliadin is a highly immunogenic protein, and B cell epitopes along its entire immunogenic length are homologous to the products of numerous proteins relevant to schizophrenia (p = 0.012 to 3e-25). These include members of the DISC1 interactome, of glutamate, dopamine and neuregulin signalling networks, and of pathways involved in plasticity, dendritic growth or myelination. Antibodies to gliadin are likely to cross react with these key proteins, as has already been observed with synapsin 1 and calreticulin. Gliadin may thus be a causative agent in schizophrenia, under certain genetic and immunological conditions, producing its effects via antibody mediated knockdown of multiple proteins relevant to the disease process. Because of such homology, an autoimmune response may be sustained by the human antigens that resemble gliadin itself, a scenario supported by many reports of immune activation both in the brain and in lymphocytes in schizophrenia. Gluten free diets and removal of such antibodies may be of therapeutic benefit in certain cases of schizophrenia. 2 Introduction A number of studies from China, Norway, and the USA have reported the presence of gliadin antibodies in schizophrenia 1-5. Gliadin is a component of gluten, intolerance to which is implicated in coeliac disease 6. -

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

Single-Cell Rnaseq Reveals Seven Classes of Colonic Sensory Neuron
Gut Online First, published on February 26, 2018 as 10.1136/gutjnl-2017-315631 Neurogastroenterology ORIGINAL ARTICLE Gut: first published as 10.1136/gutjnl-2017-315631 on 26 February 2018. Downloaded from Single-cell RNAseq reveals seven classes of colonic sensory neuron James R F Hockley,1,2 Toni S Taylor,1 Gerard Callejo,1 Anna L Wilbrey,2 Alex Gutteridge,2 Karsten Bach,1 Wendy J Winchester,2 David C Bulmer,1 Gordon McMurray,2 Ewan St John Smith1 ► Additional material is ABSTRact pathways to the central nervous system (CNS).1 In published online only. To view Objective Integration of nutritional, microbial and the colorectum, sensory innervation is organised please visit the journal online (http:// dx. doi. org/ 10. 1136/ inflammatory events along the gut-brain axis can alter into two main pathways: thoracolumbar (TL) spinal gutjnl- 2017- 315631). bowel physiology and organism behaviour. Colonic afferents projecting via the lumbar splanchnic sensory neurons activate reflex pathways and give nerve (LSN) and lumbosacral (LS) spinal afferents 1Department of Pharmacology, University of Cambridge, rise to conscious sensation, but the diversity and projecting via the pelvic nerve (PN) that are respon- Cambridge, UK division of function within these neurons is poorly sible for transducing conscious sensations of full- 2Neuroscience and Pain understood. The identification of signalling pathways ness, discomfort, urgency and pain, in addition to Research Unit, Pfizer, contributing to visceral sensation is constrained by a reflex actions.2 Cambridge, UK paucity of molecular markers. Here we address this by Visceral sensory afferents act to maintain many comprehensive transcriptomic profiling and unsupervised aspects of GI physiology, such as continence and Correspondence to James R F Hockley, Department clustering of individual mouse colonic sensory neurons. -

F2RL2 Antibody Cat
F2RL2 Antibody Cat. No.: 56-323 F2RL2 Antibody F2RL2 Antibody immunohistochemistry analysis in formalin fixed and paraffin embedded human heart tissue followed by peroxidase conjugation of the secondary antibody and DAB staining. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human This F2RL2 antibody is generated from rabbits immunized with a KLH conjugated IMMUNOGEN: synthetic peptide between 21-50 amino acids from the N-terminal region of human F2RL2. TESTED APPLICATIONS: IHC-P, WB For WB starting dilution is: 1:1000 APPLICATIONS: For IHC-P starting dilution is: 1:10~50 PREDICTED MOLECULAR 43 kDa WEIGHT: September 25, 2021 1 https://www.prosci-inc.com/f2rl2-antibody-56-323.html Properties This antibody is purified through a protein A column, followed by peptide affinity PURIFICATION: purification. CLONALITY: Polyclonal ISOTYPE: Rabbit Ig CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: Supplied in PBS with 0.09% (W/V) sodium azide. CONCENTRATION: batch dependent Store at 4˚C for three months and -20˚C, stable for up to one year. As with all antibodies STORAGE CONDITIONS: care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: F2RL2 Proteinase-activated receptor 3, PAR-3, Coagulation factor II receptor-like 2, Thrombin ALTERNATE NAMES: receptor-like 2, F2RL2, PAR3 ACCESSION NO.: O00254 GENE ID: 2151 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Coagulation factor II (thrombin) receptor-like 2 (F2RL2) is a member of the large family of 7-transmembrane-region receptors that couple to guanosine-nucleotide-binding proteins. -
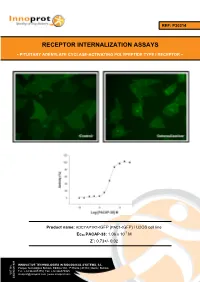
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

CXCL13/CXCR5 Interaction Facilitates VCAM-1-Dependent Migration in Human Osteosarcoma
International Journal of Molecular Sciences Article CXCL13/CXCR5 Interaction Facilitates VCAM-1-Dependent Migration in Human Osteosarcoma 1, 2,3,4, 5 6 7 Ju-Fang Liu y, Chiang-Wen Lee y, Chih-Yang Lin , Chia-Chia Chao , Tsung-Ming Chang , Chien-Kuo Han 8, Yuan-Li Huang 8, Yi-Chin Fong 9,10,* and Chih-Hsin Tang 8,11,12,* 1 School of Oral Hygiene, College of Oral Medicine, Taipei Medical University, Taipei City 11031, Taiwan; [email protected] 2 Department of Orthopaedic Surgery, Chang Gung Memorial Hospital, Puzi City, Chiayi County 61363, Taiwan; [email protected] 3 Department of Nursing, Division of Basic Medical Sciences, and Chronic Diseases and Health Promotion Research Center, Chang Gung University of Science and Technology, Puzi City, Chiayi County 61363, Taiwan 4 Research Center for Industry of Human Ecology and Research Center for Chinese Herbal Medicine, Chang Gung University of Science and Technology, Guishan Dist., Taoyuan City 33303, Taiwan 5 School of Medicine, China Medical University, Taichung 40402, Taiwan; [email protected] 6 Department of Respiratory Therapy, Fu Jen Catholic University, New Taipei City 24205, Taiwan; [email protected] 7 School of Medicine, Institute of Physiology, National Yang-Ming University, Taipei City 11221, Taiwan; [email protected] 8 Department of Biotechnology, College of Health Science, Asia University, Taichung 40402, Taiwan; [email protected] (C.-K.H.); [email protected] (Y.-L.H.) 9 Department of Sports Medicine, College of Health Care, China Medical University, Taichung 40402, Taiwan 10 Department of Orthopedic Surgery, China Medical University Beigang Hospital, Yunlin 65152, Taiwan 11 Department of Pharmacology, School of Medicine, China Medical University, Taichung 40402, Taiwan 12 Chinese Medicine Research Center, China Medical University, Taichung 40402, Taiwan * Correspondence: [email protected] (Y.-C.F.); [email protected] (C.-H.T.); Tel.: +886-4-2205-2121-7726 (C.-H.T.); Fax: +886-4-2233-3641 (C.-H.T.) These authors contributed equally to this work. -

Supplementary Data
Supplemental Data A novel mouse model of X-linked nephrogenic diabetes insipidus: Phenotypic analysis and therapeutic implications Jian Hua Li, Chung-Lin Chou, Bo Li, Oksana Gavrilova, Christoph Eisner, Jürgen Schnermann, Stasia A. Anderson, Chu-Xia Deng, Mark A. Knepper, and Jürgen Wess Supplemental Methods Metabolic cage studies. Animals were maintained in mouse metabolic cages (Hatteras Instruments, Cary, NC) under controlled temperature and light conditions (12 hr light and dark cycles). Mice received a fixed daily ration of 6.5 g of gelled diet per 20 g of body weight per day. The gelled diet was composed of 4 g of Basal Diet 5755 (Test Diet, Richmond, IN), 2.5 ml of deionized water, and 65 mg agar. Preweighted drinking water was provided ad libitum during the course of the study. Mice were acclimated in the metabolic cages for 1-2 days. Urine was collected under mineral oil in preweighted collection vials for successive 24 hr periods. Analysis of GPCR expression in mouse IMCD cells via TaqMan real-time qRT-PCR. Total RNA prepared from mouse IMCD tubule suspensions was reverse transcribed as described under Experimental Procedures. Tissues from ten 10-week old C57BL/6 WT mice were collected and pooled for each individual experiment. cDNA derived from 640 ng of RNA was mixed with an equal volume of TaqMan gene expression 2 x master mix (Applied Biosystems, Foster City, CA). 100 μl-aliquots of this mixture (corresponding to 80 ng of RNA) were added to each of the 8 fill ports of a 384-well plate of a mouse GPCR array panel (Applied Biosystems). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Original Article Expression of Chemokine Receptor CXCR5 in Gastric Cancer and Its Clinical Significance
Int J Clin Exp Pathol 2016;9(7):7202-7208 www.ijcep.com /ISSN:1936-2625/IJCEP0023559 Original Article Expression of chemokine receptor CXCR5 in gastric cancer and its clinical significance Qing Sun*, Lujun Chen*, Bin Xu, Qi Wang, Xiao Zheng, Peng Du, Dachuan Zhang, Changping Wu, Jingting Jiang Department of Tumor Biological Treatment, The Third Affiliated Hospital, Soochow University, Jiangsu Engineering Research Center for Tumor Immunotherapy, Changzhou, Jiangsu, China. *Equal contributors. Received January 8, 2016; Accepted March 22, 2016; Epub July 1, 2016; Published July 15, 2016 Abstract: The increased expression of chemokine receptor CXCR5 in cancers has been demonstrated. In order to characterize the expression pattern of CXCR5 in cell lines and tissues of gastric cancer and to assess clinical implications, the expression of CXCR5 mRNA in gastric cancer tissues and adjacent tissues was evaluated by real- time RT-PCR. Meanwhile, the expression of CXCR5 in cell lines of human gastric cancer was also analyzed by flow cytometry. Tissue microarray and immunohistochemistry were used to detect the protein expression of CXCR5 in human gastric cancer tissues and adjacent normal tissues. Flow cytometry results revealed the positive expression of CXCR5 in human gastric cancer cell lines such as BGC-823, SGC-7901 and HGC-27 cells. The immunohistochem- istry results showed higher expression of CXCR5 in 52.87% of gastric cancer tissues. The expression of CXCR5 in patients with tumor size less than 2.8 cm subgroup was significantly lower than that in patients with tumor size larger than 2.8 cm subgroup (P = 0.0456). There was no significant correlation between the expression of CXCR5 and other clinical parameters in gastric cancer. -

Flow Reagents Single Color Antibodies CD Chart
CD CHART CD N° Alternative Name CD N° Alternative Name CD N° Alternative Name Beckman Coulter Clone Beckman Coulter Clone Beckman Coulter Clone T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells CD1a T6, R4, HTA1 Act p n n p n n S l CD99 MIC2 gene product, E2 p p p CD223 LAG-3 (Lymphocyte activation gene 3) Act n Act p n CD1b R1 Act p n n p n n S CD99R restricted CD99 p p CD224 GGT (γ-glutamyl transferase) p p p p p p CD1c R7, M241 Act S n n p n n S l CD100 SEMA4D (semaphorin 4D) p Low p p p n n CD225 Leu13, interferon induced transmembrane protein 1 (IFITM1). p p p p p CD1d R3 Act S n n Low n n S Intest CD101 V7, P126 Act n p n p n n p CD226 DNAM-1, PTA-1 Act n Act Act Act n p n CD1e R2 n n n n S CD102 ICAM-2 (intercellular adhesion molecule-2) p p n p Folli p CD227 MUC1, mucin 1, episialin, PUM, PEM, EMA, DF3, H23 Act p CD2 T11; Tp50; sheep red blood cell (SRBC) receptor; LFA-2 p S n p n n l CD103 HML-1 (human mucosal lymphocytes antigen 1), integrin aE chain S n n n n n n n l CD228 Melanotransferrin (MT), p97 p p CD3 T3, CD3 complex p n n n n n n n n n l CD104 integrin b4 chain; TSP-1180 n n n n n n n p p CD229 Ly9, T-lymphocyte surface antigen p p n p n -

Molecular Characterization of Clonal Human Renal Forming Cells Cohen-Zontag Osnat , Gershon Rotem , Harari-Steinberg Orit , Kant
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.05.978254; this version posted March 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Molecular characterization of clonal human renal forming cells Cohen-Zontag Osnat1,7,*, Gershon Rotem1,7,*, Harari-Steinberg Orit1,7,*, Kanter Itamar4, Omer Dorit1,7 , Pleniceanu Oren1,7, Tam Gal4, Oriel Sarit4, Ben-Hur Herzl8,9 , Katz Guy1,3,5,7, Zohar Dotan2,7, Kalisky Tomer4,#, Dekel Benjamin1, 6,7,#,^, Pode- Shakked Naomi1,3,7,#. 1Pediatric Stem Cell Research Institute, Edmond and Lily Safra Children's Hospital, Sheba Medical Center, Tel-Hashomer, Israel 2Dept of Urology, Sheba Medical Center, Tel-Hashomer, Israel 3The Talpiot Medical Leadership Program, Sheba Medical Center, Tel-Hashomer, Israel 4Faculty of Engineering and Bar-Ilan Institute of Nanotechnology and Advanced Materials (BINA), Bar-Ilan University, Ramat Gan, Israel. 5The Joseph Buchman Gynecology and Maternity Center, Sheba Medical Center, Tel- Hashomer, Israel 6Division of Pediatric Nephrology, Edmond and Lily Safra Children's Hospital, Sheba Medical Center, Tel-Hashomer, Israel 7Sackler Faculty of Medicine, Tel-Aviv University, Tel-Aviv, Israel 8L.E.M. Laboratory of Early Detection, Nes Ziona, Israel 9Department of Obstetrics and Gynecology, Assaf Harofeh Medical Center, Tzrifin, Israel *These first authors contributed equally to this work #These senior authors contributed equally to this work ^Correspondence: Benjamin Dekel MD, PhD Pediatric Stem Cell Research Institute Edmond & Lily Safra Children's Hospital, Sheba Medical Center E-mails: [email protected] or [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.03.05.978254; this version posted March 6, 2020.