Multiplexed Analysis of the Secretin-Like GPCR-RAMP Interactome 2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -
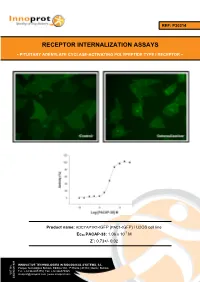
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

Secretin and Autism: a Clue but Not a Cure
SCIENCE & MEDICINE Secretin and Autism: A Clue But Not a Cure by Clarence E. Schutt, Ph.D. he world of autism has been shaken by NBC’s broadcast connections could not be found. on Dateline of a film segment documenting the effect of Tsecretin on restoring speech and sociability to autistic chil- The answer was provided nearly one hundred years ago by dren. At first blush, it seems unlikely that an intestinal hormone Bayless and Starling, who discovered that it is not nerve signals, regulating bicarbonate levels in the stomach in response to a but rather a novel substance that stimulates secretion from the good meal might influence the language centers of the brain so cells forming the intestinal mucosa. They called this substance profoundly. However, recent discoveries in neurobiology sug- “secretin.” They suggested that there could be many such cir- gest several ways of thinking about the secretin-autism connec- culating substances, or molecules, and they named them “hor- tion that could lead to the breakthroughs we dream about. mones” based on the Greek verb meaning “to excite”. As a parent with more than a decade of experience in consider- A simple analogy might help. If the body is regarded as a commu- ing a steady stream of claims of successful treatments, and as a nity of mutual service providers—the heart and muscles are the pri- scientist who believes that autism is a neurobiological disorder, I mary engines of movement, the stomach breaks down foods for have learned to temper my hopes about specific treatments by distribution, the liver detoxifies, and so on—then the need for a sys- seeing if I could construct plausible neurobiological mechanisms tem of messages conveyed by the blood becomes clear. -

Constitutive Activation of G Protein-Coupled Receptors and Diseases: Insights Into Mechanisms of Activation and Therapeutics
Pharmacology & Therapeutics 120 (2008) 129–148 Contents lists available at ScienceDirect Pharmacology & Therapeutics journal homepage: www.elsevier.com/locate/pharmthera Associate editor: S. Enna Constitutive activation of G protein-coupled receptors and diseases: Insights into mechanisms of activation and therapeutics Ya-Xiong Tao ⁎ Department of Anatomy, Physiology and Pharmacology, 212 Greene Hall, College of Veterinary Medicine, Auburn University, Auburn, AL 36849, USA article info abstract The existence of constitutive activity for G protein-coupled receptors (GPCRs) was first described in 1980s. In Keywords: 1991, the first naturally occurring constitutively active mutations in GPCRs that cause diseases were reported G protein-coupled receptor Disease in rhodopsin. Since then, numerous constitutively active mutations that cause human diseases were reported Constitutively active mutation in several additional receptors. More recently, loss of constitutive activity was postulated to also cause Inverse agonist diseases. Animal models expressing some of these mutants confirmed the roles of these mutations in the Mechanism of activation pathogenesis of the diseases. Detailed functional studies of these naturally occurring mutations, combined Transgenic model with homology modeling using rhodopsin crystal structure as the template, lead to important insights into the mechanism of activation in the absence of crystal structure of GPCRs in active state. Search for inverse Abbreviations: agonists on these receptors will be critical for correcting the diseases cause by activating mutations in GPCRs. ADRP, autosomal dominant retinitis pigmentosa Theoretically, these inverse agonists are better therapeutics than neutral antagonists in treating genetic AgRP, Agouti-related protein AR, adrenergic receptor diseases caused by constitutively activating mutations in GPCRs. CAM, constitutively active mutant © 2008 Elsevier Inc. -

Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes
Tohoku J. Exp. Med., 2011,Differential 223, 161-176 Gene Expression in OPCs, Oligodendrocytes and Type II Astrocytes 161 Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes Jian-Guo Hu,1,2,* Yan-Xia Wang,3,* Jian-Sheng Zhou,2 Chang-Jie Chen,4 Feng-Chao Wang,1 Xing-Wu Li1 and He-Zuo Lü1,2 1Department of Clinical Laboratory Science, The First Affiliated Hospital of Bengbu Medical College, Bengbu, P.R. China 2Anhui Key Laboratory of Tissue Transplantation, Bengbu Medical College, Bengbu, P.R. China 3Department of Neurobiology, Shanghai Jiaotong University School of Medicine, Shanghai, P.R. China 4Department of Laboratory Medicine, Bengbu Medical College, Bengbu, P.R. China Oligodendrocyte precursor cells (OPCs) are bipotential progenitor cells that can differentiate into myelin-forming oligodendrocytes or functionally undetermined type II astrocytes. Transplantation of OPCs is an attractive therapy for demyelinating diseases. However, due to their bipotential differentiation potential, the majority of OPCs differentiate into astrocytes at transplanted sites. It is therefore important to understand the molecular mechanisms that regulate the transition from OPCs to oligodendrocytes or astrocytes. In this study, we isolated OPCs from the spinal cords of rat embryos (16 days old) and induced them to differentiate into oligodendrocytes or type II astrocytes in the absence or presence of 10% fetal bovine serum, respectively. RNAs were extracted from each cell population and hybridized to GeneChip with 28,700 rat genes. Using the criterion of fold change > 4 in the expression level, we identified 83 genes that were up-regulated and 89 genes that were down-regulated in oligodendrocytes, and 92 genes that were up-regulated and 86 that were down-regulated in type II astrocytes compared with OPCs. -

Insights Into Nuclear G-Protein-Coupled Receptors As Therapeutic Targets in Non-Communicable Diseases
pharmaceuticals Review Insights into Nuclear G-Protein-Coupled Receptors as Therapeutic Targets in Non-Communicable Diseases Salomé Gonçalves-Monteiro 1,2, Rita Ribeiro-Oliveira 1,2, Maria Sofia Vieira-Rocha 1,2, Martin Vojtek 1,2 , Joana B. Sousa 1,2,* and Carmen Diniz 1,2,* 1 Laboratory of Pharmacology, Department of Drug Sciences, Faculty of Pharmacy, University of Porto, 4050-313 Porto, Portugal; [email protected] (S.G.-M.); [email protected] (R.R.-O.); [email protected] (M.S.V.-R.); [email protected] (M.V.) 2 LAQV/REQUIMTE, Faculty of Pharmacy, University of Porto, 4050-313 Porto, Portugal * Correspondence: [email protected] (J.B.S.); [email protected] (C.D.) Abstract: G-protein-coupled receptors (GPCRs) comprise a large protein superfamily divided into six classes, rhodopsin-like (A), secretin receptor family (B), metabotropic glutamate (C), fungal mating pheromone receptors (D), cyclic AMP receptors (E) and frizzled (F). Until recently, GPCRs signaling was thought to emanate exclusively from the plasma membrane as a response to extracellular stimuli but several studies have challenged this view demonstrating that GPCRs can be present in intracellular localizations, including in the nuclei. A renewed interest in GPCR receptors’ superfamily emerged and intensive research occurred over recent decades, particularly regarding class A GPCRs, but some class B and C have also been explored. Nuclear GPCRs proved to be functional and capable of triggering identical and/or distinct signaling pathways associated with their counterparts on the cell surface bringing new insights into the relevance of nuclear GPCRs and highlighting the Citation: Gonçalves-Monteiro, S.; nucleus as an autonomous signaling organelle (triggered by GPCRs). -
![RT² Profiler PCR Array (96-Well Format and 384-Well [4 X 96] Format)](https://docslib.b-cdn.net/cover/5806/rt%C2%B2-profiler-pcr-array-96-well-format-and-384-well-4-x-96-format-905806.webp)
RT² Profiler PCR Array (96-Well Format and 384-Well [4 X 96] Format)
RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human GPCR Signaling PathwayFinder Cat. no. 330231 PAHS-071ZA For pathway expression analysis Format For use with the following real-time cyclers RT² Profiler PCR Array, Applied Biosystems® models 5700, 7000, 7300, 7500, Format A 7700, 7900HT, ViiA™ 7 (96-well block); Bio-Rad® models iCycler®, iQ™5, MyiQ™, MyiQ2; Bio-Rad/MJ Research Chromo4™; Eppendorf® Mastercycler® ep realplex models 2, 2s, 4, 4s; Stratagene® models Mx3005P®, Mx3000P®; Takara TP-800 RT² Profiler PCR Array, Applied Biosystems models 7500 (Fast block), 7900HT (Fast Format C block), StepOnePlus™, ViiA 7 (Fast block) RT² Profiler PCR Array, Bio-Rad CFX96™; Bio-Rad/MJ Research models DNA Format D Engine Opticon®, DNA Engine Opticon 2; Stratagene Mx4000® RT² Profiler PCR Array, Applied Biosystems models 7900HT (384-well block), ViiA 7 Format E (384-well block); Bio-Rad CFX384™ RT² Profiler PCR Array, Roche® LightCycler® 480 (96-well block) Format F RT² Profiler PCR Array, Roche LightCycler 480 (384-well block) Format G RT² Profiler PCR Array, Fluidigm® BioMark™ Format H Sample & Assay Technologies Description The Human G-Protein-Coupled Receptor Signaling PathwayFinder RT² Profiler PCR Array profiles the expression of 84 genes representative of and involved in GPCR-mediated signal transduction pathways. G-protein Coupled Receptors (GPCRs) that are represented on this array include bioactive lipid receptors, metabotropic glutamate receptors (mGluRs), GABA-B-like receptors, rhodopsin-like receptors, and secretin-like receptors. Members of the intricate network of intracellular signaling pathways involved in GPCR-mediated signal transduction are also represented on this array including the cAMP / PKA Pathway, Calcium / PKC Pathway, Calcium / NFAT Pathway, PLC Pathway, Protein Tyrosine Kinase Pathway, PKC / MEK Pathway, p43 / p44MAP Pathway, p38 MAP Pathway, PI-3 Kinase Pathway, NO-cGMP Pathway, Rho Pathway, NFκB Pathway and Jak / Stat Pathway. -

Single-Cell Analysis of Crohn's Disease Lesions Identifies
bioRxiv preprint doi: https://doi.org/10.1101/503102; this version posted December 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy JC Martin1,2,3, G Boschetti1,2,3, C Chang1,2,3, R Ungaro4, M Giri5, LS Chuang5, S Nayar5, A Greenstein6, M. Dubinsky7, L Walker1,2,5,8, A Leader1,2,3, JS Fine9, CE Whitehurst9, L Mbow9, S Kugathasan10, L.A. Denson11, J.Hyams12, JR Friedman13, P Desai13, HM Ko14, I Laface1,2,8, Guray Akturk1,2,8, EE Schadt15,16, S Gnjatic1,2,8, A Rahman1,2,5,8, , M Merad1,2,3,8,17,18*, JH Cho5,17,*, E Kenigsberg1,15,16,17* 1 Precision Immunology Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 2 Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 3 Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 4 The Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York City, NY 10029, USA. 5 Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 6 Department of Colorectal Surgery, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA 7 Department of Pediatrics, Susan and Leonard Feinstein IBD Clinical Center, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Multi-Functionality of Proteins Involved in GPCR and G Protein Signaling: Making Sense of Structure–Function Continuum with In
Cellular and Molecular Life Sciences (2019) 76:4461–4492 https://doi.org/10.1007/s00018-019-03276-1 Cellular andMolecular Life Sciences REVIEW Multi‑functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder‑based proteoforms Alexander V. Fonin1 · April L. Darling2 · Irina M. Kuznetsova1 · Konstantin K. Turoverov1,3 · Vladimir N. Uversky2,4 Received: 5 August 2019 / Revised: 5 August 2019 / Accepted: 12 August 2019 / Published online: 19 August 2019 © Springer Nature Switzerland AG 2019 Abstract GPCR–G protein signaling system recognizes a multitude of extracellular ligands and triggers a variety of intracellular signal- ing cascades in response. In humans, this system includes more than 800 various GPCRs and a large set of heterotrimeric G proteins. Complexity of this system goes far beyond a multitude of pair-wise ligand–GPCR and GPCR–G protein interactions. In fact, one GPCR can recognize more than one extracellular signal and interact with more than one G protein. Furthermore, one ligand can activate more than one GPCR, and multiple GPCRs can couple to the same G protein. This defnes an intricate multifunctionality of this important signaling system. Here, we show that the multifunctionality of GPCR–G protein system represents an illustrative example of the protein structure–function continuum, where structures of the involved proteins represent a complex mosaic of diferently folded regions (foldons, non-foldons, unfoldons, semi-foldons, and inducible foldons). The functionality of resulting highly dynamic conformational ensembles is fne-tuned by various post-translational modifcations and alternative splicing, and such ensembles can undergo dramatic changes at interaction with their specifc partners. -

Immunobiology of VPAC2 Receptor
Aus der Medizinischen Poliklinik-Innenstadt der Ludwig-Maximilians-Universität München Direktor: Professor Dr. med. M. Reincke Immunobiology of the VPAC2 Receptor Dissertation zum Erwerb des Doktorgrades der Medizin an der Medizinischen Fakultät der Ludwig-Maximilians-Universität zu München Vorgelegt von Carola Grinninger aus München 2010 Mit Genehmigung der Medizinischen Fakultät der Universität München Berichterstatter: Prof. Dr. med. S. Schewe Mitberichterstatter: PD Dr. B. Bachmeier Prof. Dr. H. Kellner Prof. Dr. U. Gresser Betreuung durch den promovierten Mitarbeiter: PD Dr. P. J. Nelson Dekan: Prof. Dr. med. Dr. h.c. M. Reiser, FACR, FRCR Tag der mündlichen Prüfung: 07.10.2010 2 CONTENTS Contents……………………………………………………………………….…3 Summary…………………………………………………………………………5 1. Introduction……………………………………………………………………10 1.1 General introduction……………………………………………………...10 1.2 G-Protein-coupled Receptors…………………………………………….10 1.3 G-protein-coupled signalling…………………………………………….13 1.4 Molecularbiology and structure of VPAC Receptors………………...….15 1.5 Vasoactive Intestinal Peptide (VIP)……………………………………...22 1.5.1 Biochemical structure of vasoactive intestinal peptide………...............22 1.6 VIP and intracellular signaling…………………………………………...25 1.7 VIP and rheumatoid arthritis…………………………………..…………26 1.8 Hypothesis………………………………………………………………..29 2. Materials……………………………………………………..………………31 2.1 Bacteria ……………………………………………………………….…31 2.2 Vectors……………………………………………………………….…..31 2.3 Cells………………………………………………………………….…..32 2.4 Mice……………………………………………………………….….….33 2.5 Oligonucleotides………………………………………………….….…..33