Vassilatis Et Al Suppl
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Development and Maintenance of Epidermal Stem Cells in Skin Adnexa
International Journal of Molecular Sciences Review Development and Maintenance of Epidermal Stem Cells in Skin Adnexa Jaroslav Mokry * and Rishikaysh Pisal Medical Faculty, Charles University, 500 03 Hradec Kralove, Czech Republic; [email protected] * Correspondence: [email protected] Received: 30 October 2020; Accepted: 18 December 2020; Published: 20 December 2020 Abstract: The skin surface is modified by numerous appendages. These structures arise from epithelial stem cells (SCs) through the induction of epidermal placodes as a result of local signalling interplay with mesenchymal cells based on the Wnt–(Dkk4)–Eda–Shh cascade. Slight modifications of the cascade, with the participation of antagonistic signalling, decide whether multipotent epidermal SCs develop in interfollicular epidermis, scales, hair/feather follicles, nails or skin glands. This review describes the roles of epidermal SCs in the development of skin adnexa and interfollicular epidermis, as well as their maintenance. Each skin structure arises from distinct pools of epidermal SCs that are harboured in specific but different niches that control SC behaviour. Such relationships explain differences in marker and gene expression patterns between particular SC subsets. The activity of well-compartmentalized epidermal SCs is orchestrated with that of other skin cells not only along the hair cycle but also in the course of skin regeneration following injury. This review highlights several membrane markers, cytoplasmic proteins and transcription factors associated with epidermal SCs. Keywords: stem cell; epidermal placode; skin adnexa; signalling; hair pigmentation; markers; keratins 1. Epidermal Stem Cells as Units of Development 1.1. Development of the Epidermis and Placode Formation The embryonic skin at very early stages of development is covered by a surface ectoderm that is a precursor to the epidermis and its multiple derivatives. -

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

Single-Cell Rnaseq Reveals Seven Classes of Colonic Sensory Neuron
Gut Online First, published on February 26, 2018 as 10.1136/gutjnl-2017-315631 Neurogastroenterology ORIGINAL ARTICLE Gut: first published as 10.1136/gutjnl-2017-315631 on 26 February 2018. Downloaded from Single-cell RNAseq reveals seven classes of colonic sensory neuron James R F Hockley,1,2 Toni S Taylor,1 Gerard Callejo,1 Anna L Wilbrey,2 Alex Gutteridge,2 Karsten Bach,1 Wendy J Winchester,2 David C Bulmer,1 Gordon McMurray,2 Ewan St John Smith1 ► Additional material is ABSTRact pathways to the central nervous system (CNS).1 In published online only. To view Objective Integration of nutritional, microbial and the colorectum, sensory innervation is organised please visit the journal online (http:// dx. doi. org/ 10. 1136/ inflammatory events along the gut-brain axis can alter into two main pathways: thoracolumbar (TL) spinal gutjnl- 2017- 315631). bowel physiology and organism behaviour. Colonic afferents projecting via the lumbar splanchnic sensory neurons activate reflex pathways and give nerve (LSN) and lumbosacral (LS) spinal afferents 1Department of Pharmacology, University of Cambridge, rise to conscious sensation, but the diversity and projecting via the pelvic nerve (PN) that are respon- Cambridge, UK division of function within these neurons is poorly sible for transducing conscious sensations of full- 2Neuroscience and Pain understood. The identification of signalling pathways ness, discomfort, urgency and pain, in addition to Research Unit, Pfizer, contributing to visceral sensation is constrained by a reflex actions.2 Cambridge, UK paucity of molecular markers. Here we address this by Visceral sensory afferents act to maintain many comprehensive transcriptomic profiling and unsupervised aspects of GI physiology, such as continence and Correspondence to James R F Hockley, Department clustering of individual mouse colonic sensory neurons. -

F2RL2 Antibody Cat
F2RL2 Antibody Cat. No.: 56-323 F2RL2 Antibody F2RL2 Antibody immunohistochemistry analysis in formalin fixed and paraffin embedded human heart tissue followed by peroxidase conjugation of the secondary antibody and DAB staining. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human This F2RL2 antibody is generated from rabbits immunized with a KLH conjugated IMMUNOGEN: synthetic peptide between 21-50 amino acids from the N-terminal region of human F2RL2. TESTED APPLICATIONS: IHC-P, WB For WB starting dilution is: 1:1000 APPLICATIONS: For IHC-P starting dilution is: 1:10~50 PREDICTED MOLECULAR 43 kDa WEIGHT: September 25, 2021 1 https://www.prosci-inc.com/f2rl2-antibody-56-323.html Properties This antibody is purified through a protein A column, followed by peptide affinity PURIFICATION: purification. CLONALITY: Polyclonal ISOTYPE: Rabbit Ig CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: Supplied in PBS with 0.09% (W/V) sodium azide. CONCENTRATION: batch dependent Store at 4˚C for three months and -20˚C, stable for up to one year. As with all antibodies STORAGE CONDITIONS: care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: F2RL2 Proteinase-activated receptor 3, PAR-3, Coagulation factor II receptor-like 2, Thrombin ALTERNATE NAMES: receptor-like 2, F2RL2, PAR3 ACCESSION NO.: O00254 GENE ID: 2151 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Coagulation factor II (thrombin) receptor-like 2 (F2RL2) is a member of the large family of 7-transmembrane-region receptors that couple to guanosine-nucleotide-binding proteins. -

Mechanical Forces Induce an Asthma Gene Signature in Healthy Airway Epithelial Cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T
www.nature.com/scientificreports OPEN Mechanical forces induce an asthma gene signature in healthy airway epithelial cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T. Kho4, Kelan Tantisira1, Marc Santolini 1,5, Feixiong Cheng6,7,8, Jennifer A. Mitchel3, Maureen McGill3, Michael J. O’Sullivan3, Margherita De Marzio1,3, Amitabh Sharma1, Scott H. Randell9, Jefrey M. Drazen3, Jefrey J. Fredberg3 & Scott T. Weiss1,3* Bronchospasm compresses the bronchial epithelium, and this compressive stress has been implicated in asthma pathogenesis. However, the molecular mechanisms by which this compressive stress alters pathways relevant to disease are not well understood. Using air-liquid interface cultures of primary human bronchial epithelial cells derived from non-asthmatic donors and asthmatic donors, we applied a compressive stress and then used a network approach to map resulting changes in the molecular interactome. In cells from non-asthmatic donors, compression by itself was sufcient to induce infammatory, late repair, and fbrotic pathways. Remarkably, this molecular profle of non-asthmatic cells after compression recapitulated the profle of asthmatic cells before compression. Together, these results show that even in the absence of any infammatory stimulus, mechanical compression alone is sufcient to induce an asthma-like molecular signature. Bronchial epithelial cells (BECs) form a physical barrier that protects pulmonary airways from inhaled irritants and invading pathogens1,2. Moreover, environmental stimuli such as allergens, pollutants and viruses can induce constriction of the airways3 and thereby expose the bronchial epithelium to compressive mechanical stress. In BECs, this compressive stress induces structural, biophysical, as well as molecular changes4,5, that interact with nearby mesenchyme6 to cause epithelial layer unjamming1, shedding of soluble factors, production of matrix proteins, and activation matrix modifying enzymes, which then act to coordinate infammatory and remodeling processes4,7–10. -
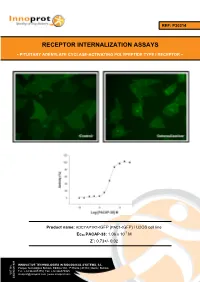
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

CD157 and CD200 at the Crossroads of Endothelial Remodeling and Immune Regulation
Editorial Commentary Page 1 of 7 CD157 and CD200 at the crossroads of endothelial remodeling and immune regulation Masuko Katoh1, Masaru Katoh2 1M & M PrecMed, Tokyo, Japan; 2Department of Omics Network, National Cancer Center, Tokyo, Japan Correspondence to: Masaru Katoh. Department of Omics Network, National Cancer Center, 5-1-1 Tsukiji, Chuo-ward, Tokyo 104-0045, Japan. Email: [email protected]. Comment on: Wakabayashi T, Naito H, Suehiro JI, et al. CD157 Marks Tissue-Resident Endothelial Stem Cells with Homeostatic and Regenerative Properties. Cell Stem Cell 2018;22:384-97.e6. Received: 11 March 2019; Accepted: 08 April 2019; Published: 19 April 2019. doi: 10.21037/sci.2019.04.01 View this article at: http://dx.doi.org/10.21037/sci.2019.04.01 The endothelial cells that coat the inner wall of blood cells formed more CD31 (PECAM1)-positive colonies than vessels are essential for the maintenance of the vascular CD200 single-positive or CD157/CD200 double-negative network, metabolic homeostasis and stem cell populations liver endothelial cells in vitro. The expression levels of Atf3, in tissue or tumor microenvironments (1-3). Angiogenesis Fosl2, Myc and Sox7 were significantly upregulated in the is defined as neovascular formation through the sprouting CD157/CD200 double-positive cells compared with the and proliferation of endothelial cells from preexisting CD200 single-positive or CD157/CD200 double-negative blood vessels. VEGF (VEGFA) and FGFs that transduce cells; however, the functions of these transcription factors signals through VEGFR2 and FGFRs, respectively, are in CD157/CD200 double-positive endothelial cells remain representative proangiogenic factors (4,5). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcriptomic Analysis of Native Versus Cultured Human and Mouse Dorsal Root Ganglia Focused on Pharmacological Targets Short
bioRxiv preprint doi: https://doi.org/10.1101/766865; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Transcriptomic analysis of native versus cultured human and mouse dorsal root ganglia focused on pharmacological targets Short title: Comparative transcriptomics of acutely dissected versus cultured DRGs Andi Wangzhou1, Lisa A. McIlvried2, Candler Paige1, Paulino Barragan-Iglesias1, Carolyn A. Guzman1, Gregory Dussor1, Pradipta R. Ray1,#, Robert W. Gereau IV2, # and Theodore J. Price1, # 1The University of Texas at Dallas, School of Behavioral and Brain Sciences and Center for Advanced Pain Studies, 800 W Campbell Rd. Richardson, TX, 75080, USA 2Washington University Pain Center and Department of Anesthesiology, Washington University School of Medicine # corresponding authors [email protected], [email protected] and [email protected] Funding: NIH grants T32DA007261 (LM); NS065926 and NS102161 (TJP); NS106953 and NS042595 (RWG). The authors declare no conflicts of interest Author Contributions Conceived of the Project: PRR, RWG IV and TJP Performed Experiments: AW, LAM, CP, PB-I Supervised Experiments: GD, RWG IV, TJP Analyzed Data: AW, LAM, CP, CAG, PRR Supervised Bioinformatics Analysis: PRR Drew Figures: AW, PRR Wrote and Edited Manuscript: AW, LAM, CP, GD, PRR, RWG IV, TJP All authors approved the final version of the manuscript. 1 bioRxiv preprint doi: https://doi.org/10.1101/766865; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Celsr1-3 Cadherins in PCP and Brain Development
CHAPTER SEVEN Celsr1–3 Cadherins in PCP and Brain Development Camille Boutin, André M. Goffinet1, Fadel Tissir1 Institute of Neuroscience, Developmental Neurobiology, Universite´ Catholique de Louvain, Brussels, Belgium 1Corresponding authors: Equal contribution. e-mail address: [email protected]; andre. [email protected] Contents 1. Celsr1–3 Expression Patterns 164 2. Celsr1: A Major Player in Vertebrate PCP 165 3. Celsr2 and 3 in Ciliogenesis 169 4. Celsr1–3 in Neuronal Migration 171 5. Celsr2 and Celsr3 in Brain Wiring 174 5.1 Motifs of Celsr important for their functions 176 References 179 Abstract Cadherin EGF LAG seven-pass G-type receptors 1, 2, and 3 (Celsr1–3) form a family of three atypical cadherins with multiple functions in epithelia and in the nervous system. During the past decade, evidence has accumulated for important and distinct roles of Celsr1–3 in planar cell polarity (PCP) and brain development and maintenance. Although the role of Celsr in PCP is conserved from flies to mammals, other functions may be more distantly related, with Celsr working only with one or a subset of the classical PCP partners. Here, we review the literature on Celsr in PCP and neural devel- opment, point to several remaining questions, and consider future challenges and possible research trends. Celsr1–3 genes encode atypical cadherins of more than 3000 amino acids ( Fig. 7.1). Their large ectodomain is composed of nine N-terminal cadherin repeats (typical cadherins have five repeats), six epidermal growth factor (EGF)-like domains, two laminin G repeats, one hormone receptor motif (HRM), and a G-protein-coupled receptor proteolytic site (GPS). -

Flow Reagents Single Color Antibodies CD Chart
CD CHART CD N° Alternative Name CD N° Alternative Name CD N° Alternative Name Beckman Coulter Clone Beckman Coulter Clone Beckman Coulter Clone T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells CD1a T6, R4, HTA1 Act p n n p n n S l CD99 MIC2 gene product, E2 p p p CD223 LAG-3 (Lymphocyte activation gene 3) Act n Act p n CD1b R1 Act p n n p n n S CD99R restricted CD99 p p CD224 GGT (γ-glutamyl transferase) p p p p p p CD1c R7, M241 Act S n n p n n S l CD100 SEMA4D (semaphorin 4D) p Low p p p n n CD225 Leu13, interferon induced transmembrane protein 1 (IFITM1). p p p p p CD1d R3 Act S n n Low n n S Intest CD101 V7, P126 Act n p n p n n p CD226 DNAM-1, PTA-1 Act n Act Act Act n p n CD1e R2 n n n n S CD102 ICAM-2 (intercellular adhesion molecule-2) p p n p Folli p CD227 MUC1, mucin 1, episialin, PUM, PEM, EMA, DF3, H23 Act p CD2 T11; Tp50; sheep red blood cell (SRBC) receptor; LFA-2 p S n p n n l CD103 HML-1 (human mucosal lymphocytes antigen 1), integrin aE chain S n n n n n n n l CD228 Melanotransferrin (MT), p97 p p CD3 T3, CD3 complex p n n n n n n n n n l CD104 integrin b4 chain; TSP-1180 n n n n n n n p p CD229 Ly9, T-lymphocyte surface antigen p p n p n -

In-Depth Characterization of the Wnt-Signaling/Β-Catenin Pathway In
Götzel et al. BMC Gastroenterology (2019) 19:38 https://doi.org/10.1186/s12876-019-0957-5 RESEARCH ARTICLE Open Access In-depth characterization of the Wnt- signaling/β-catenin pathway in an in vitro model of Barrett’s sequence Katharina Götzel1, Olga Chemnitzer1, Luisa Maurer1, Arne Dietrich1,2, Uwe Eichfeld1, Orestis Lyros1, Yusef Moulla1, Stefan Niebisch1, Matthias Mehdorn1, Boris Jansen-Winkeln1, Michael Vieth3, Albrecht Hoffmeister4, Ines Gockel1 and René Thieme1* Abstract Background: An altered Wnt-signaling activation has been reported during Barrett’s esophagus progression, but with rarely detected mutations in APC and β-catenin (CTNNB1) genes. Methods: In this study, a robust in-depth expression pattern analysis of frizzled receptors, co-receptors, the Wnt- ligands Wnt3a and Wnt5a, the Wnt-signaling downstream targets Axin2, and CyclinD1, as well as the activation of the intracellular signaling kinases Akt and GSK3β was performed in an in vitro cell culture model of Barrett’s esophagus. Representing the Barrett’s sequence, we used normal esophageal squamous epithelium (EPC-1, EPC-2), metaplasia (CP-A) and dysplasia (CP-B) to esophageal adenocarcinoma (EAC) cell lines (OE33, OE19) and primary specimens of squamous epithelium, metaplasia and EAC. Results: A loss of Wnt3a expression was observed beginning from the metaplastic cell line CP-A towards dysplasia (CP-B) and EAC (OE33 and OE19), confirmed by a lower staining index of WNT3A in Barrett’s metaplasia and EAC, than in squamous epithelium specimens. Frizzled 1–10 expression analysis revealed a distinct expression pattern, showing the highest expression for Fzd2, Fzd3, Fzd4, Fzd5, Fzd7, and the co-receptor LRP5/6 in EAC cells, while Fzd3 and Fzd7 were rarely expressed in primary specimens from squamous epithelium.