SUPPLEMENTARY APPENDIX Targetable Driver Mutations in Multicentric Reticulohistiocytosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

Genetic Associations Between Voltage-Gated Calcium Channels (Vgccs) and Autism Spectrum Disorder (ASD)
Liao and Li Molecular Brain (2020) 13:96 https://doi.org/10.1186/s13041-020-00634-0 REVIEW Open Access Genetic associations between voltage- gated calcium channels and autism spectrum disorder: a systematic review Xiaoli Liao1,2 and Yamin Li2* Abstract Objectives: The present review systematically summarized existing publications regarding the genetic associations between voltage-gated calcium channels (VGCCs) and autism spectrum disorder (ASD). Methods: A comprehensive literature search was conducted to gather pertinent studies in three online databases. Two authors independently screened the included records based on the selection criteria. Discrepancies in each step were settled through discussions. Results: From 1163 resulting searched articles, 28 were identified for inclusion. The most prominent among the VGCCs variants found in ASD were those falling within loci encoding the α subunits, CACNA1A, CACNA1B, CACN A1C, CACNA1D, CACNA1E, CACNA1F, CACNA1G, CACNA1H, and CACNA1I as well as those of their accessory subunits CACNB2, CACNA2D3, and CACNA2D4. Two signaling pathways, the IP3-Ca2+ pathway and the MAPK pathway, were identified as scaffolds that united genetic lesions into a consensus etiology of ASD. Conclusions: Evidence generated from this review supports the role of VGCC genetic variants in the pathogenesis of ASD, making it a promising therapeutic target. Future research should focus on the specific mechanism that connects VGCC genetic variants to the complex ASD phenotype. Keywords: Autism spectrum disorder, Voltage-gated calcium -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

C1orf21 CRISPR/Cas9 KO Plasmid (H): Sc-417269
SANTA CRUZ BIOTECHNOLOGY, INC. C1orf21 CRISPR/Cas9 KO Plasmid (h): sc-417269 BACKGROUND APPLICATIONS The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and C1orf21 CRISPR/Cas9 KO Plasmid (h) is recommended for the disruption of CRISPR-associated protein (Cas9) system is an adaptive immune response gene expression in human cells. defense mechanism used by archea and bacteria for the degradation of foreign genetic material (4,6). This mechanism can be repurposed for other 20 nt non-coding RNA sequence: guides Cas9 functions, including genomic engineering for mammalian systems, such as to a specific target location in the genomic DNA gene knockout (KO) (1,2,3,5). CRISPR/Cas9 KO Plasmid products enable the U6 promoter: drives gRNA scaffold: helps Cas9 identification and cleavage of specific genes by utilizing guide RNA (gRNA) expression of gRNA bind to target DNA sequences derived from the Genome-scale CRISPR Knock-Out (GeCKO) v2 library developed in the Zhang Laboratory at the Broad Institute (3,5). Termination signal Green Fluorescent Protein: to visually REFERENCES verify transfection CRISPR/Cas9 Knockout Plasmid CBh (chicken β-Actin 1. Cong, L., et al. 2013. Multiplex genome engineering using CRISPR/Cas hybrid) promoter: drives expression of Cas9 systems. Science 339: 819-823. 2A peptide: allows production of both Cas9 and GFP from the 2. Mali, P., et al. 2013. RNA-guided human genome engineering via Cas9. same CBh promoter Science 339: 823-826. Nuclear localization signal 3. Ran, F.A., et al. 2013. Genome engineering using the CRISPR-Cas9 system. Nuclear localization signal SpCas9 ribonuclease Nat. Protoc. 8: 2281-2308. -

The Mineralocorticoid Receptor Leads to Increased Expression of EGFR
www.nature.com/scientificreports OPEN The mineralocorticoid receptor leads to increased expression of EGFR and T‑type calcium channels that support HL‑1 cell hypertrophy Katharina Stroedecke1,2, Sandra Meinel1,2, Fritz Markwardt1, Udo Kloeckner1, Nicole Straetz1, Katja Quarch1, Barbara Schreier1, Michael Kopf1, Michael Gekle1 & Claudia Grossmann1* The EGF receptor (EGFR) has been extensively studied in tumor biology and recently a role in cardiovascular pathophysiology was suggested. The mineralocorticoid receptor (MR) is an important efector of the renin–angiotensin–aldosterone‑system and elicits pathophysiological efects in the cardiovascular system; however, the underlying molecular mechanisms are unclear. Our aim was to investigate the importance of EGFR for MR‑mediated cardiovascular pathophysiology because MR is known to induce EGFR expression. We identifed a SNP within the EGFR promoter that modulates MR‑induced EGFR expression. In RNA‑sequencing and qPCR experiments in heart tissue of EGFR KO and WT mice, changes in EGFR abundance led to diferential expression of cardiac ion channels, especially of the T‑type calcium channel CACNA1H. Accordingly, CACNA1H expression was increased in WT mice after in vivo MR activation by aldosterone but not in respective EGFR KO mice. Aldosterone‑ and EGF‑responsiveness of CACNA1H expression was confrmed in HL‑1 cells by Western blot and by measuring peak current density of T‑type calcium channels. Aldosterone‑induced CACNA1H protein expression could be abrogated by the EGFR inhibitor AG1478. Furthermore, inhibition of T‑type calcium channels with mibefradil or ML218 reduced diameter, volume and BNP levels in HL‑1 cells. In conclusion the MR regulates EGFR and CACNA1H expression, which has an efect on HL‑1 cell diameter, and the extent of this regulation seems to depend on the SNP‑216 (G/T) genotype. -
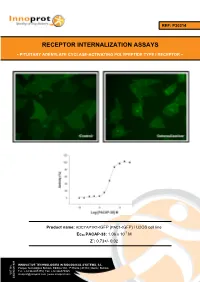
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

An Interactive Web Application to Explore Regeneration-Associated Gene Expression and Chromatin Accessibility
Supplementary Materials Regeneration Rosetta: An interactive web application to explore regeneration-associated gene expression and chromatin accessibility Andrea Rau, Sumona P. Dhara, Ava J. Udvadia, Paul L. Auer 1. Table S1. List of cholesterol metabolic genes from MGI database 2. Table S2. List of differentially expressed transcripts during optic nerve regeneration in zebrafish using the MGI cholesterol metabolic gene queries in the Regeneration Rosetta app 3. Table S3. List of transcription factor encoding genes from brain cell bodies following spinal cord injury in lamprey over a course of 12 weeKs 4. Table S4. List of transcription factor encoding genes from spinal cell bodies following spinal cord injury in lamprey over a course of 12 weeks Ensembl ID MGI Gene ID Symbol Name ENSMUSG00000015243 MGI:99607 Abca1 ATP-binding cassette, sub-family A (ABC1), member 1 ENSMUSG00000026944 MGI:99606 Abca2 ATP-binding cassette, sub-family A (ABC1), member 2 ENSMUSG00000024030 MGI:107704 Abcg1 ATP binding cassette subfamily G member 1 ENSMUSG00000026003 MGI:87866 Acadl acyl-Coenzyme A dehydrogenase, long-chain ENSMUSG00000018574 MGI:895149 Acadvl acyl-Coenzyme A dehydrogenase, very long chain ENSMUSG00000038641 MGI:2384785 Akr1d1 aldo-keto reductase family 1, member D1 ENSMUSG00000028553 MGI:1353627 Angptl3 angiopoietin-like 3 ENSMUSG00000031996 MGI:88047 Aplp2 amyloid beta (A4) precursor-like protein 2 ENSMUSG00000032083 MGI:88049 Apoa1 apolipoprotein A-I ENSMUSG00000005681 MGI:88050 Apoa2 apolipoprotein A-II ENSMUSG00000032080 MGI:88051 Apoa4 -

Aquaporin Channels in the Heart—Physiology and Pathophysiology
International Journal of Molecular Sciences Review Aquaporin Channels in the Heart—Physiology and Pathophysiology Arie O. Verkerk 1,2,* , Elisabeth M. Lodder 2 and Ronald Wilders 1 1 Department of Medical Biology, Amsterdam University Medical Centers, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands; [email protected] 2 Department of Experimental Cardiology, Amsterdam University Medical Centers, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands; [email protected] * Correspondence: [email protected]; Tel.: +31-20-5664670 Received: 29 March 2019; Accepted: 23 April 2019; Published: 25 April 2019 Abstract: Mammalian aquaporins (AQPs) are transmembrane channels expressed in a large variety of cells and tissues throughout the body. They are known as water channels, but they also facilitate the transport of small solutes, gasses, and monovalent cations. To date, 13 different AQPs, encoded by the genes AQP0–AQP12, have been identified in mammals, which regulate various important biological functions in kidney, brain, lung, digestive system, eye, and skin. Consequently, dysfunction of AQPs is involved in a wide variety of disorders. AQPs are also present in the heart, even with a specific distribution pattern in cardiomyocytes, but whether their presence is essential for proper (electro)physiological cardiac function has not intensively been studied. This review summarizes recent findings and highlights the involvement of AQPs in normal and pathological cardiac function. We conclude that AQPs are at least implicated in proper cardiac water homeostasis and energy balance as well as heart failure and arsenic cardiotoxicity. However, this review also demonstrates that many effects of cardiac AQPs, especially on excitation-contraction coupling processes, are virtually unexplored. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Palmitoyl-Protein Thioesterase 1 Deficiency in Drosophila Melanogaster Causes Accumulation
Genetics: Published Articles Ahead of Print, published on February 1, 2006 as 10.1534/genetics.105.053306 Palmitoyl-protein thioesterase 1 deficiency in Drosophila melanogaster causes accumulation of abnormal storage material and reduced lifespan Anthony J. Hickey*,†,1, Heather L. Chotkowski*, Navjot Singh*, Jeffrey G. Ault*, Christopher A. Korey‡,2, Marcy E. MacDonald‡, and Robert L. Glaser*,†,3 * Wadsworth Center, New York State Department of Health, Albany, NY 12201-2002 † Department of Biomedical Sciences, State University of New York, Albany, NY 12201-0509 ‡ Molecular Neurogenetics Unit, Center for Human Genetic Research, Massachusetts General Hospital, Boston, MA 02114 1 current address: Albany Medical College, Albany, NY 12208 2 current address: Department of Biology, College of Charleston, Charleston, SC 294243 3 corresponding author: Wadsworth Center, NYS Dept. Health, P. O. Box 22002, Albany, NY 12201-2002 E-mail: [email protected] 1 running title: Phenotypes of Ppt1-deficient Drosophila key words: Batten disease infantile neuronal ceroid lipofuscinosis palmitoyl-protein thioesterase CLN1 Drosophila corresponding author: Robert L. Glaser Wadsworth Center, NYS Dept. Health P. O. Box 22002 Albany, NY 12201-2002 E-mail: [email protected] phone: 518-473-4201 fax: 518-474-3181 2 ABSTRACT Human neuronal ceroid lipofuscinoses (NCLs) are a group of genetic neurodegenerative diseases characterized by progressive death of neurons in the central nervous system (CNS) and accumulation of abnormal lysosomal storage material. Infantile NCL (INCL), the most severe form of NCL, is caused by mutations in the Ppt1 gene, which encodes the lysosomal enzyme palmitoyl-protein thioesterase 1 (Ppt1). We generated mutations in the Ppt1 ortholog of Drosophila melanogaster in order to characterize phenotypes caused by Ppt1-deficiency in flies. -

Investigation of Copy Number Variations on Chromosome 21 Detected by Comparative Genomic Hybridization
Li et al. Molecular Cytogenetics (2018) 11:42 https://doi.org/10.1186/s13039-018-0391-3 RESEARCH Open Access Investigation of copy number variations on chromosome 21 detected by comparative genomic hybridization (CGH) microarray in patients with congenital anomalies Wenfu Li, Xianfu Wang and Shibo Li* Abstract Background: The clinical features of Down syndrome vary among individuals, with those most common being congenital heart disease, intellectual disability, developmental abnormity and dysmorphic features. Complex combination of Down syndrome phenotype could be produced by partially copy number variations (CNVs) on chromosome 21 as well. By comparing individual with partial CNVs of chromosome 21 with other patients of known CNVs and clinical phenotypes, we hope to provide a better understanding of the genotype-phenotype correlation of chromosome 21. Methods: A total of 2768 pediatric patients sample collected at the Genetics Laboratory at Oklahoma University Health Science Center were screened using CGH Microarray for CNVs on chromosome 21. Results: We report comprehensive clinical and molecular descriptions of six patients with microduplication and seven patients with microdeletion on the long arm of chromosome 21. Patients with microduplication have varied clinical features including developmental delay, microcephaly, facial dysmorphic features, pulmonary stenosis, autism, preauricular skin tag, eye pterygium, speech delay and pain insensitivity. We found that patients with microdeletion presented with developmental delay, microcephaly, intrauterine fetal demise, epilepsia partialis continua, congenital coronary anomaly and seizures. Conclusion: Three patients from our study combine with four patients in public database suggests an association between 21q21.1 microduplication of CXADR gene and patients with developmental delay. One patient with 21q22.13 microdeletion of DYRK1A shows association with microcephaly and scoliosis. -
![Cd1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719](https://docslib.b-cdn.net/cover/1446/cd1a-o10-concentrated-and-prediluted-monoclonal-antibody-901-3158-061719-341446.webp)
Cd1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719
CD1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719 Catalog Number: ACI 3158 A, B API 3158 AA VLTM 3158 G20 Description: 0.1, 0.5 mL conc. 6.0 mL, RTU 20 mL, RTU Dilution: 1:100 Ready-to-use Ready-to-use Diluent: Van Gogh Yellow N/A N/A Intended Use: Protocol Recommendations (VALENT® Automated Slide For In Vitro Diagnostic Use Staining Platform) Cont’d: CD1a [O10] is a mouse monoclonal antibody that is intended for Protein Block (Optional): Incubate for 10-20 minutes at RT with Val laboratory use in the qualitative identification of CD1a protein by Background Block. immunohistochemistry (IHC) in formalin-fixed paraffin-embedded Primary Antibody: Incubate for 30 minutes. (FFPE) human tissues. The clinical interpretation of any staining or its Secondary: Incubate for 10 minutes with Val Mouse Secondary. absence should be complemented by morphological studies using proper Linker: Incubate for 10 minutes with Val Universal Linker. controls and should be evaluated within the context of the patient’s Polymer: Incubate for 10 minutes with Val Universal Polymer. clinical history and other diagnostic tests by a qualified pathologist. Chromogen: Incubate for 5 minutes with Val DAB. Summary and Explanation: Counterstain: Counterstain for 5 minutes with Val Hematoxylin. CD1a is a protein of 43 to 49 kDa and is expressed on dendritic cells and cortical thymocytes (1,2). CD1a [O10] staining has been shown to be Protocol Recommendations (intelliPATH FLX® and manual use): useful in the differentiation of Langerhans cells from interdigitating cells. Peroxide Block: Block for 5 minutes with Peroxidazed 1. It has also proved useful for phenotyping Langerhans cell histiocytosis Pretreatment: Perform heat retrieval using Diva or Reveal Decloaker.