Peripheral Nerve Single-Cell Analysis Identifies Mesenchymal Ligands That Promote Axonal Growth
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

Epha4/Tie2 Crosstalk Regulates Leptomeningeal Collateral Remodeling Following Ischemic Stroke
EphA4/Tie2 crosstalk regulates leptomeningeal collateral remodeling following ischemic stroke Benjamin Okyere, … , John B. Matson, Michelle H. Theus J Clin Invest. 2019. https://doi.org/10.1172/JCI131493. Research In-Press Preview Neuroscience Vascular biology Leptomeningeal anastomoses or pial collateral vessels play a critical role in cerebral blood flow (CBF) restoration following ischemic stroke. The magnitude of this adaptive response is postulated to be controlled by the endothelium, although the underlying molecular mechanisms remain under investigation. Here we demonstrated that endothelial genetic deletion, using EphA4f/f/Tie2-Cre and EphA4f/f/VeCahderin-CreERT2 mice and vessel painting strategies, implicated EphA4 receptor tyrosine kinase as a major suppressor of pial collateral remodeling, CBF and functional recovery following permanent middle cerebral artery occlusion. Pial collateral remodeling is limited by the cross talk between EphA4-Tie2 signaling in vascular endothelial cells, which is mediated through p-Akt regulation. Furthermore, peptide inhibition of EphA4 resulted in acceleration of the pial arteriogenic response. Our findings demonstrate EphA4 is a negative regulator of Tie2 receptor signaling which limits pial collateral arteriogenesis following cerebrovascular occlusion. Therapeutic targeting of EphA4 and/or Tie2 represents an attractive new strategy for improving collateral function, neural tissue health and functional recovery following ischemic stroke. Find the latest version: https://jci.me/131493/pdf 1 EphA4/Tie2 -
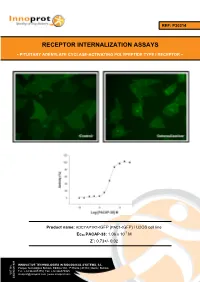
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Structural Characterization of Polysaccharides from Cordyceps Militaris and Their Hypolipidemic Effects Cite This: RSC Adv.,2018,8,41012 in High Fat Diet Fed Mice†
RSC Advances View Article Online PAPER View Journal | View Issue Structural characterization of polysaccharides from Cordyceps militaris and their hypolipidemic effects Cite this: RSC Adv.,2018,8,41012 in high fat diet fed mice† Zhen-feng Huang, ‡ Ming-long Zhang,‡ Song Zhang,* Ya-hui Wang and Xue-wen Jiang Cordyceps militaris is a crude dietary therapeutic mushroom with high nutritional and medicinal values. Mushroom-derived polysaccharides have been found to possess antihyperglycemic and antihyperlipidemic activities. This study aimed to partially clarify the structural characterization and comparatively evaluate hypolipidemic potentials of intracellular- (IPCM) and extracellular polysaccharides of C. militaris (EPCM) in high fat diet fed mice. Results indicated that IPCM-2 is a-pyran polysaccharide with an average molecular weight of 32.5 kDa, was mainly composed of mannose, glucose and galactose with mass percentages of 51.94%, 10.54%, and 37.25%, respectively. EPCM-2 is an a-pyran Creative Commons Attribution 3.0 Unported Licence. polysaccharide with an average molecular weight of 20 kDa that is mainly composed of mannose, glucose and galactose with mass percentages of 44.51%, 18.33%, and 35.38%, respectively. In in vivo study, EPCM-1 treatment (100 mg kgÀ1 dÀ1) showed potential effects on improving serum lipid profiles of hyperlipidemic mice, reflected by decreasing serum total cholesterol (TC), triglyceride (TG) and low density lipoprotein-cholesterol (LDL-C) levels by 20.05%, 45.45% and 52.63%, respectively, while IPCM-1 treatment -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Purkinje Cell Migration Disorder By
CEREBELLAR CORTICOGENESIS IN THE LYSOSOMAL ACID PHOSPHATASE (ACP2) MUTANT MICE: PURKINJE CELL MIGRATION DISORDER BY NILOUFAR ASHTARI A Thesis Submitted to the Faculty of Graduate Studies of The University of Manitoba in Partial Fulfilment of the Requirements for the Degree of MASTER OF SCIENCE Department of Human Anatomy and Cell Science University of Manitoba Winnipeg, Manitoba Copyright © 2017 by Niloufar Ashtari 1 Abstract In a mutant mouse called nax as the result of mutation in Lysosomal Acid phosphatase (Acp2), layers of the cerebellar cortex are impaired and monolayer Purkinje cells (Pcs) turn to multi-layered Pcs that ectopically invade the molecular layer. We investigated reelin-Dab1 signaling as an important pathway for Pcs migration and monolayer formation in cerebellum. ERK1/2 is a member of mitogen activated kinases family and suggested to be a downstream of reelin signaling. We hypothesize that the establishment of mono-layered Pcs rely on reelin through ERK1/2 pathway. Acp2 mutant mice were used for this study and molecular expression and distribution were assessed by immunohistochemistry, RT-PCR, western blotting, and cell culture. Results suggest that reelin may modulate the ERK1/2 expression, thus lower expression of reelin and higher phosphorylation of Dab1 leads to over expression of the ERK1/2 that causes the Pcs to over migrate and form multilayer in nax cerebellar cortex. i TABLE OF CONTENTS LISTOFABBREVIATIONS………………………………………………..…... IV LIST OF TABLES……………………………………...…………………...….. Vii LIST OF FIGURES…………………………………………………….………. Viii CHAPTER 1: INTRODUCTION…………………………………….………… 1 1.1 Cerebellum ……………………………………………………........………. 1 1.2 Development of Central Nervous System………………………………….. 2 1.3 Development of the cerebellum………………………………….................. 3 1.4 Specification of cerebellar germinal zones…………………………………. -

Gene Expression Analysis Identifies Potential Biomarkers of Neurofibromatosis Type 1 Including Adrenomedullin
Published OnlineFirst August 25, 2010; DOI: 10.1158/1078-0432.CCR-10-0613 Clinical Imaging, Diagnosis, Prognosis Cancer Research Gene Expression Analysis Identifies Potential Biomarkers of Neurofibromatosis Type 1 Including Adrenomedullin Trent R. Hummel1, Walter J. Jessen1, Shyra J. Miller1, Lan Kluwe4, Victor F. Mautner4, Margaret R. Wallace5, Conxi Lázaro6, Grier P. Page7, Paul F. Worley8, Bruce J. Aronow2, Elizabeth K. Schorry3, and Nancy Ratner1 Abstract Purpose: Plexiform neurofibromas (pNF) are Schwann cell tumors found in a third of individuals with neurofibromatosis type 1 (NF1). pNF can undergo transformation to malignant peripheral nerve sheath tumors (MPNST). There are no identified serum biomarkers of pNF tumor burden or transformation to MPNST. Serum biomarkers would be useful to verify NF1 diagnosis, monitor tumor burden, and/or detect transformation. Experimental Design: We used microarray gene expression analysis to define 92 genes that encode putative secreted proteins in neurofibroma Schwann cells, neurofibromas, and MPNST. We validated dif- ferential expression by quantitative reverse transcription-PCR, Western blotting, and ELISA assays in cell conditioned medium and control and NF1 patient sera. Results: Of 13 candidate genes evaluated, only adrenomedullin (ADM) was confirmed as differentially expressed and elevated in serum of NF1 patients. ADM protein concentrati on was further elevated in serum of a small sampling of NF1 patients with MPNST. MPNST cell conditioned medium, containing ADM and hepatocyte growth factor, stimulated MPNST migration and endothelial cell proliferation. Conclusions: Thus, microarray analysis identifies potential serum biomarkers for disease, and ADM is a serum biomarker of NF1. ADM serum levels do not seem to correlate with the presence of pNFs but may be a biomarker of transformation to MPNST. -

Characterization of Pulmonary Arteriovenous Malformations in ACVRL1 Versus ENG Mutation Carriers in Hereditary Hemorrhagic Telangiectasia
© American College of Medical Genetics and Genomics ORIGINAL RESEARCH ARTICLE Characterization of pulmonary arteriovenous malformations in ACVRL1 versus ENG mutation carriers in hereditary hemorrhagic telangiectasia Weiyi Mu, ScM1, Zachary A. Cordner, MD, PhD2, Kevin Yuqi Wang, MD3, Kate Reed, MPH, ScM4, Gina Robinson, RN5, Sally Mitchell, MD5 and Doris Lin, MD, PhD5 Purpose: Pulmonary arteriovenous malformations (pAVMs) are mutation carriers to have pAVMs (P o 0.001) or multiple lesions major contributors to morbidity and mortality in hereditary (P = 0.03), and to undergo procedural intervention (P = 0.02). hemorrhagic telangiectasia (HHT). Mutations in ENG and ACVRL1 Additionally, pAVMs in ENG carriers were more likely to exhibit underlie the vast majority of clinically diagnosed cases. The aims of bilateral lung involvement and growth over time, although this did this study were to characterize and compare the clinical and not reach statistical significance. The HHT severity score was morphologic features of pAVMs between these two genotype significantly higher in ENG than in ACVRL1 (P = 0.02). groups. Conclusion: The propensity and multiplicity of ENG-associated Methods: Sixty-six patients with HHT and affected family pAVMs may contribute to the higher disease severity in this members were included. Genotype, phenotypic data, and imaging genotype, as reflected by the HHT severity score and the frequency were obtained from medical records. Morphologic features of of interventional procedures. pAVMs were analyzed using computed tomography angiography. Genet Med HHT symptoms, pAVM imaging characteristics, frequency of advance online publication 19 October 2017 procedural intervention, and HHT severity scores were compared Key Words: ACVRL1; ENG; genotype-phenotype correlation; between ENG and ACVRL1 genotype groups. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

Genotyping of Breech Flystrike Resource – Update
Project No: ON-00515 Contract No: PO4500010753 AWI Project Manager: Bridget Peachey Contractor Name: CSIRO Agriculture and Food Prepared by: Dr Sonja Dominik Publication Date: July 2019 Genotyping of breech flystrike resource – update Published by Australian Wool Innovation Limited, Level 6, 68 Harrington Street, THE ROCKS, NSW, 2000 This publication should only be used as a general aid and is not a substitute for specific advice. To the extent permitted by law, we exclude all liability for loss or damage arising from the use of the information in this publication. AWI invests in research, development, innovation and marketing activities along the global supply chain for Australian wool. AWI is grateful for its funding, which is primarily provided by Australian woolgrowers through a wool levy and by the Australian Government which provides a matching contribution for eligible R&D activities © 2019 Australian Wool Innovation Ltd. All rights reserved. Contents Executive Summary .................................................................................................................... 3 1 Introduction/Hypothesis .................................................................................................... 5 2 Literature Review ............................................................................................................... 6 3 Project Objectives .............................................................................................................. 8 4 Success in Achieving Objectives ........................................................................................ -

3 Cleavage Products of Notch 2/Site and Myelopoiesis by Dysregulating
ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch This information is current as David R. Gibb, Sheinei J. Saleem, Dae-Joong Kang, Mark of October 4, 2021. A. Subler and Daniel H. Conrad J Immunol 2011; 186:4244-4252; Prepublished online 2 March 2011; doi: 10.4049/jimmunol.1003318 http://www.jimmunol.org/content/186/7/4244 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2011/03/02/jimmunol.100331 Material 8.DC1 http://www.jimmunol.org/ References This article cites 45 articles, 16 of which you can access for free at: http://www.jimmunol.org/content/186/7/4244.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 4, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch David R. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated.