Supporting Online Material
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Comparative Gene Expression Profiling of Stromal Cell Matrices
ell Res C ea m rc te h S & f o T h l Journal of Tiwari et al., J Stem Cell Res Ther 2013, 3:4 e a r n a r p u DOI: 10.4172/2157-7633.1000152 y o J ISSN: 2157-7633 Stem Cell Research & Therapy Research Article Open Access Comparative Gene Expression Profiling of Stromal Cell Matrices that Support Expansion of Hematopoietic Stem/Progenitor Cells Abhilasha Tiwari1,2, Christophe Lefevre2, Mark A Kirkland2*, Kevin Nicholas2 and Gopal Pande1* 1CSIR-Centre for Cellular and Molecular Biology (CCMB), Hyderabad, India 2Deakin University, Waurn Ponds, Geelong, VIC, Australia Abstract The bone marrow microenvironment maintains a stable balance between self-renewal and differentiation of hematopoietic stem/progenitor cells (HSPCs). This microenvironment, also termed the “hematopoietic niche”, is primarily composed of stromal cells and their extracellular matrices (ECM) that jointly regulate HSPC functions. Previously, we have demonstrated that umbilical cord blood derived HSPCs can be maintained and expanded on stromal cell derived acellular matrices that mimic the complexity of the hematopoietic niche. The results indicated that matrices prepared at 20% O2 with osteogenic medium (OGM) were best suited for expanding committed HSPCs, whereas, matrices prepared at 5% O2 without OGM were better for primitive progenitors. Based upon these results we proposed that individual constituents of these matrices could be responsible for regulation of specific HSPC functions. To explore this hypothesis, we have performed comparative transcriptome profiling of these matrix producing cells, which identified differential expression of both known niche regulators, such as Wnt4, Angpt2, Vcam and Cxcl12, as well as genes not previously associated with HSPC regulation, such as Depp. -

HDAC7 Sikens the Heart
The black sheep of class IIa: HDAC7 SIKens the heart Joshua G. Travers, … , Tianjing Hu, Timothy A. McKinsey J Clin Invest. 2020;130(6):2811-2813. https://doi.org/10.1172/JCI137074. Commentary Class IIa histone deacetylases (HDACs) repress cardiomyocyte hypertrophy through association with the prohypertrophic transcription factor (TF) myocyte enhancer factor-2 (MEF2). The four class IIa HDACs — HDAC4, -5, -7, and -9 — are subject to signal-dependent phosphorylation by members of the Ca2+/calmodulin-dependent protein kinase (CaMK) group. In response to stress, HDAC4, HDAC5, and HDAC9 undergo phosphorylation-induced nuclear export in cardiomyocytes, freeing MEF2 to stimulate progrowth genes; it was generally assumed that HDAC7 is also antihypertrophic. However, in this issue of the JCI, Hsu and colleagues demonstrate that, in sharp contrast to the other class IIa HDACs, HDAC7 is constitutively localized to the cardiomyocyte cytoplasm, where it promotes cardiac hypertrophy. Phosphorylation of HDAC7 by the CaMK group member salt-inducible kinase 1 (SIK1) stabilized the deacetylase, leading to increased expression of c-Myc, which in turn stimulated a pathological gene program. These unexpected findings highlight the SIK1/HDAC7 signaling axis as a promising target for the treatment of cardiac hypertrophy and heart failure. Find the latest version: https://jci.me/137074/pdf The Journal of Clinical Investigation COMMENTARY The black sheep of class IIa: HDAC7 SIKens the heart Joshua G. Travers, Tianjing Hu, and Timothy A. McKinsey Division of Cardiology, Department of Medicine, and Consortium for Fibrosis Research & Translation, University of Colorado Anschutz Medical Campus, Aurora, Colorado, USA. repress gene expression, block cardiac hypertrophy by associating with the myo- Class IIa histone deacetylases (HDACs) repress cardiomyocyte hypertrophy cyte enhancer factor-2 (MEF2) transcrip- through association with the prohypertrophic transcription factor (TF) tion factor (TF) (11). -

Differential Gene Expression Profile in Endometrioid And
[CANCER RESEARCH 63, 5697–5702, September 15, 2003] Advances in Brief Differential Gene Expression Profile in Endometrioid and Nonendometrioid Endometrial Carcinoma: STK15 Is Frequently Overexpressed and Amplified in Nonendometrioid Carcinomas1 Gema Moreno-Bueno, Carolina Sa´nchez-Este´vez, Rau´l Cassia, Sandra Rodrı´guez-Perales, Ramo´n Dı´az-Uriarte, Orlando Domı´nguez, David Hardisson, Miguel Andujar, Jaime Prat, Xavier Matias-Guiu, Juan C. Cigudosa, and Jose´Palacios2 Laboratory of Breast and Gynaecological Cancer, Molecular Pathology Programme [G. M-B., C. S-E., R. C., J. Pa.] and Biotechnology Programme [S. R-P., R. D-U., O. D., J. C. C.], Centro Nacional de Investigaciones Oncologicas, Madrid; Department of Pathology, Hospital Universitario La Paz, Madrid [D. H.]; Department of Pathology, Hospital Materno Infantil, Las Palmas [M. A.]; Department of Pathology, Hospital Sant Pau y Sant Creu, Barcelona [J. Pr.]; and Department of Pathology, Hospital Arnau de Villanova, Lleida [X. M-G.], Spain Abstract tiated endometrioid carcinomas that usually develop in pre- and perimenopausal women. They are associated with estrogen stimula- Endometrial carcinoma (EC) comprises at least two types of cancer: tion, coexist with, or are preceded by atypical endometrial hyperplasia endometrioid carcinomas (EECs) are estrogen-related tumors, which are and are associated with ER positivity and with K-RAS, PTEN, and frequently euploid and have a good prognosis. Nonendometrioid carcino-  mas (NEECs; serous and clear cell forms) are not estrogen related, are -catenin mutations, and microsatellite instability. Conversely, type II frequently aneuploid, and are clinically aggressive. We used cDNA mi- tumors are NEECs (papillary serous and clear cell carcinomas) that croarrays containing 6386 different genes to analyze gene expression occur in older women. -

An Overview of the Role of Hdacs in Cancer Immunotherapy
International Journal of Molecular Sciences Review Immunoepigenetics Combination Therapies: An Overview of the Role of HDACs in Cancer Immunotherapy Debarati Banik, Sara Moufarrij and Alejandro Villagra * Department of Biochemistry and Molecular Medicine, School of Medicine and Health Sciences, The George Washington University, 800 22nd St NW, Suite 8880, Washington, DC 20052, USA; [email protected] (D.B.); [email protected] (S.M.) * Correspondence: [email protected]; Tel.: +(202)-994-9547 Received: 22 March 2019; Accepted: 28 April 2019; Published: 7 May 2019 Abstract: Long-standing efforts to identify the multifaceted roles of histone deacetylase inhibitors (HDACis) have positioned these agents as promising drug candidates in combatting cancer, autoimmune, neurodegenerative, and infectious diseases. The same has also encouraged the evaluation of multiple HDACi candidates in preclinical studies in cancer and other diseases as well as the FDA-approval towards clinical use for specific agents. In this review, we have discussed how the efficacy of immunotherapy can be leveraged by combining it with HDACis. We have also included a brief overview of the classification of HDACis as well as their various roles in physiological and pathophysiological scenarios to target key cellular processes promoting the initiation, establishment, and progression of cancer. Given the critical role of the tumor microenvironment (TME) towards the outcome of anticancer therapies, we have also discussed the effect of HDACis on different components of the TME. We then have gradually progressed into examples of specific pan-HDACis, class I HDACi, and selective HDACis that either have been incorporated into clinical trials or show promising preclinical effects for future consideration. -

Characterization of BRCA1-Deficient Premalignant Tissues and Cancers Identifies Plekha5 As a Tumor Metastasis Suppressor
ARTICLE https://doi.org/10.1038/s41467-020-18637-9 OPEN Characterization of BRCA1-deficient premalignant tissues and cancers identifies Plekha5 as a tumor metastasis suppressor Jianlin Liu1,2, Ragini Adhav1,2, Kai Miao1,2, Sek Man Su1,2, Lihua Mo1,2, Un In Chan1,2, Xin Zhang1,2, Jun Xu1,2, Jianjie Li1,2, Xiaodong Shu1,2, Jianming Zeng 1,2, Xu Zhang1,2, Xueying Lyu1,2, Lakhansing Pardeshi1,3, ✉ ✉ Kaeling Tan1,3, Heng Sun1,2, Koon Ho Wong 1,3, Chuxia Deng 1,2 & Xiaoling Xu 1,2 1234567890():,; Single-cell whole-exome sequencing (scWES) is a powerful approach for deciphering intra- tumor heterogeneity and identifying cancer drivers. So far, however, simultaneous analysis of single nucleotide variants (SNVs) and copy number variations (CNVs) of a single cell has been challenging. By analyzing SNVs and CNVs simultaneously in bulk and single cells of premalignant tissues and tumors from mouse and human BRCA1-associated breast cancers, we discover an evolution process through which the tumors initiate from cells with SNVs affecting driver genes in the premalignant stage and malignantly progress later via CNVs acquired in chromosome regions with cancer driver genes. These events occur randomly and hit many putative cancer drivers besides p53 to generate unique genetic and pathological features for each tumor. Upon this, we finally identify a tumor metastasis suppressor Plekha5, whose deficiency promotes cancer metastasis to the liver and/or lung. 1 Cancer Centre, Faculty of Health Sciences, University of Macau, Macau, SAR, China. 2 Centre for Precision Medicine Research and Training, Faculty of Health Sciences, University of Macau, Macau, SAR, China. -

ACE2 Interaction Networks in COVID-19: a Physiological Framework for Prediction of Outcome in Patients with Cardiovascular Risk Factors
Journal of Clinical Medicine Article ACE2 Interaction Networks in COVID-19: A Physiological Framework for Prediction of Outcome in Patients with Cardiovascular Risk Factors Zofia Wicik 1,2 , Ceren Eyileten 2, Daniel Jakubik 2,Sérgio N. Simões 3, David C. Martins Jr. 1, Rodrigo Pavão 1, Jolanta M. Siller-Matula 2,4,* and Marek Postula 2 1 Centro de Matemática, Computação e Cognição, Universidade Federal do ABC, Santo Andre 09606-045, Brazil; zofi[email protected] (Z.W.); [email protected] (D.C.M.J.); [email protected] (R.P.) 2 Department of Experimental and Clinical Pharmacology, Medical University of Warsaw, Center for Preclinical Research and Technology CEPT, 02-091 Warsaw, Poland; [email protected] (C.E.); [email protected] (D.J.); [email protected] (M.P.) 3 Federal Institute of Education, Science and Technology of Espírito Santo, Serra, Espírito Santo 29056-264, Brazil; [email protected] 4 Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, 1090 Vienna, Austria * Correspondence: [email protected]; Tel.: +43-1-40400-46140; Fax: +43-1-40400-42160 Received: 9 October 2020; Accepted: 17 November 2020; Published: 21 November 2020 Abstract: Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (coronavirus disease 2019; COVID-19) is associated with adverse outcomes in patients with cardiovascular disease (CVD). The aim of the study was to characterize the interaction between SARS-CoV-2 and Angiotensin-Converting Enzyme 2 (ACE2) functional networks with a focus on CVD. Methods: Using the network medicine approach and publicly available datasets, we investigated ACE2 tissue expression and described ACE2 interaction networks that could be affected by SARS-CoV-2 infection in the heart, lungs and nervous system. -

Dickkopf-1 Promotes Hematopoietic Regeneration Via Direct and Niche-Mediated Mechanisms
ARTICLES Dickkopf-1 promotes hematopoietic regeneration via direct and niche-mediated mechanisms Heather A Himburg1,7, Phuong L Doan2,7, Mamle Quarmyne1,3, Xiao Yan1,3, Joshua Sasine1, Liman Zhao1, Grace V Hancock4, Jenny Kan1, Katherine A Pohl1, Evelyn Tran1, Nelson J Chao2, Jeffrey R Harris2 & John P Chute1,5,6 The role of osteolineage cells in regulating hematopoietic stem cell (HSC) regeneration following myelosuppression is not well understood. Here we show that deletion of the pro-apoptotic genes Bak and Bax in osterix (Osx, also known as Sp7 transcription factor 7)-expressing cells in mice promotes HSC regeneration and hematopoietic radioprotection following total body irradiation. These mice showed increased bone marrow (BM) levels of the protein dickkopf-1 (Dkk1), which was produced in Osx-expressing BM cells. Treatment of irradiated HSCs with Dkk1 in vitro increased the recovery of both long-term repopulating HSCs and progenitor cells, and systemic administration of Dkk1 to irradiated mice increased hematopoietic recovery and improved survival. Conversely, inducible deletion of one allele of Dkk1 in Osx-expressing cells in adult mice inhibited the recovery of BM stem and progenitor cells and of complete blood counts following irradiation. Dkk1 promoted hematopoietic regeneration via both direct effects on HSCs, in which treatment with Dkk1 decreased the levels of mitochondrial reactive oxygen species and suppressed senescence, and indirect effects on BM endothelial cells, in which treatment with Dkk1 induced epidermal growth factor (EGF) secretion. Accordingly, blockade of the EGF receptor partially abrogated Dkk1-mediated hematopoietic recovery. These data identify Dkk1 as a regulator of hematopoietic regeneration and demonstrate paracrine cross-talk between BM osteolineage cells and endothelial cells in regulating hematopoietic reconstitution following injury. -

Searching for Novel Peptide Hormones in the Human Genome Olivier Mirabeau
Searching for novel peptide hormones in the human genome Olivier Mirabeau To cite this version: Olivier Mirabeau. Searching for novel peptide hormones in the human genome. Life Sciences [q-bio]. Université Montpellier II - Sciences et Techniques du Languedoc, 2008. English. tel-00340710 HAL Id: tel-00340710 https://tel.archives-ouvertes.fr/tel-00340710 Submitted on 21 Nov 2008 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITE MONTPELLIER II SCIENCES ET TECHNIQUES DU LANGUEDOC THESE pour obtenir le grade de DOCTEUR DE L'UNIVERSITE MONTPELLIER II Discipline : Biologie Informatique Ecole Doctorale : Sciences chimiques et biologiques pour la santé Formation doctorale : Biologie-Santé Recherche de nouvelles hormones peptidiques codées par le génome humain par Olivier Mirabeau présentée et soutenue publiquement le 30 janvier 2008 JURY M. Hubert Vaudry Rapporteur M. Jean-Philippe Vert Rapporteur Mme Nadia Rosenthal Examinatrice M. Jean Martinez Président M. Olivier Gascuel Directeur M. Cornelius Gross Examinateur Résumé Résumé Cette thèse porte sur la découverte de gènes humains non caractérisés codant pour des précurseurs à hormones peptidiques. Les hormones peptidiques (PH) ont un rôle important dans la plupart des processus physiologiques du corps humain. -

Plasma Omentin Levels Are Inversely Associated with Atherosclerosis In
Nishimura et al. Cardiovasc Diabetol (2019) 18:167 https://doi.org/10.1186/s12933-019-0973-3 Cardiovascular Diabetology ORIGINAL INVESTIGATION Open Access Plasma omentin levels are inversely associated with atherosclerosis in type 2 diabetes patients with increased plasma adiponectin levels: a cross-sectional study Masami Nishimura1, Tomoaki Morioka1* , Mariko Hayashi1, Yoshinori Kakutani1, Yuko Yamazaki1, Masafumi Kurajoh1, Katsuhito Mori2, Shinya Fukumoto3, Atsushi Shioi4,5, Tetsuo Shoji4,5, Masaaki Inaba1,5 and Masanori Emoto1 Abstract Background: Omentin and adiponectin are among the anti-infammatory and anti-atherogenic adipokines that have potentially benefcial efects on cardiovascular disorders. Recent studies indicate a paradoxical relationship between adiponectin and cardiovascular mortality across many clinical settings including type 2 diabetes. In this study, we characterized the clinical features of type 2 diabetes patients with increased adiponectin levels and exam- ined the association between omentin and atherosclerosis in those patients. Methods: The subjects were 413 patients with type 2 diabetes. Fasting plasma omentin and total adiponectin levels were measured by enzyme-linked immunosorbent assay. The intima-media thickness (IMT) of the common carotid artery was measured by ultrasonography. The subjects were stratifed according to the median value of plasma adiponectin. Results: In high-adiponectin group, omentin levels were higher, while IMT tended to be greater than those in low- adiponectin group. The high-adiponectin group also exhibited older age, higher systolic blood pressure, lower kidney function, body mass index, and insulin resistance index compared to the low-adiponectin group. Multivariate analysis revealed that omentin levels were independently and negatively associated with IMT in high-adiponectin group, but not in low-adiponectin group, after adjusting for adiponectin levels and traditional cardiovascular risk factors. -

(Title of the Thesis)*
THE PHYSIOLOGICAL ACTIONS OF ADIPONECTIN IN CENTRAL AUTONOMIC NUCLEI: IMPLICATIONS FOR THE INTEGRATIVE CONTROL OF ENERGY HOMEOSTASIS by Ted Donald Hoyda A thesis submitted to the Department of Physiology In conformity with the requirements for the degree of Doctor of Philosophy Queen‟s University Kingston, Ontario, Canada (September, 2009) Copyright © Ted Donald Hoyda, 2009 ABSTRACT Adiponectin regulates feeding behavior, energy expenditure and autonomic function through the activation of two receptors present in nuclei throughout the central nervous system, however much remains unknown about the mechanisms mediating these effects. Here I investigate the actions of adiponectin in autonomic centers of the hypothalamus (the paraventricular nucleus) and brainstem (the nucleus of the solitary tract) through examining molecular, electrical, hormonal and physiological consequences of peptidergic signalling. RT-PCR and in situ hybridization experiments demonstrate the presence of AdipoR1 and AdipoR2 mRNA in the paraventricular nucleus. Investigation of the electrical consequences following receptor activation in the paraventricular nucleus indicates that magnocellular-oxytocin cells are homogeneously inhibited while magnocellular-vasopressin neurons display mixed responses. Single cell RT-PCR analysis shows oxytocin neurons express both receptors while vasopressin neurons express either both receptors or one receptor. Co-expressing oxytocin and vasopressin neurons express neither receptor and are not affected by adiponectin. Median eminence projecting corticotropin releasing hormone neurons, brainstem projecting oxytocin neurons, and thyrotropin releasing hormone neurons are all depolarized by adiponectin. Plasma adrenocorticotropin hormone concentration is increased following intracerebroventricular injections of adiponectin. I demonstrate that the nucleus of the solitary tract, the primary cardiovascular regulation site of the medulla, expresses mRNA for AdipoR1 and AdipoR2 and mediates adiponectin induced hypotension. -
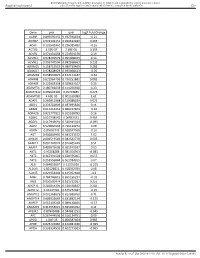
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

NIH Public Access Author Manuscript Rev Endocr Metab Disord
NIH Public Access Author Manuscript Rev Endocr Metab Disord. Author manuscript; available in PMC 2014 September 03. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Rev Endocr Metab Disord. 2014 June ; 15(2): 137–147. doi:10.1007/s11154-013-9280-6. ADIPONECTIN SIGNALING IN THE LIVER Terry P. Combs1,2,3 and Errol B. Marliss2 1Department of Medicine, University of North Carolina at Chapel Hill 2Crabtree Nutrition Laboratories, Department of Medicine, McGill University Abstract High glucose production contributes to fed and fasted hyperglycemia in Type 1 Diabetes (T1D) and Type 2 Diabetes (T2D). The breakdown of the adiponectin signaling pathway in T1D and the reduction of circulating adiponectin in T2D contribute to this abnormal increase in glucose production. Sufficient amounts of insulin could compensate for the loss of adiponectin signaling in T1D and T2D and reduce hyperglycemia. However, the combination of low adiponectin signaling and high insulin resembles an insulin resistance state associated with cardiovascular disease and decreased life expectancy. Future development of medications that correct the deficiency of adiponectin signaling in the liver could restore the metabolic balance in T1D and T2D and reduce the need for insulin. This article reviews the adiponectin signaling pathway in the liver through T- cadherin, AdipoR1, AdipoR2, AMPK, ceramidase activity, APPL1 and the recently discovered Suppressor Of Glucose from Autophagy (SOGA). 1. Liver contribution to circulating glucose The liver releases glucose ensuring sufficient amounts of this essential fuel are always available in the circulation for other tissues. Endocrine and neural mechanisms stimulate the liver during fasting, exercise and pregnancy to meet the increased demand for glucose.