Decreased Expression of BNC1 and BNC2 Is Associated with Genetic Or Epigenetic Regulation in Hepatocellular Carcinoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
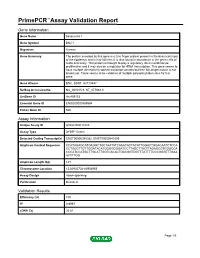
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name basonuclin 1 Gene Symbol BNC1 Organism Human Gene Summary The protein encoded by this gene is a zinc finger protein present in the basal cell layer of the epidermis and in hair follicles. It is also found in abundance in the germ cells of testis and ovary. This protein is thought to play a regulatory role in keratinocyte proliferation and it may also be a regulator for rRNA transcription. This gene seems to have multiple alternatively spliced transcript variants but their full-length nature is not known yet. There seems to be evidence of multiple polyadenylation sites for this gene. Gene Aliases BNC, BSN1, HsT19447 RefSeq Accession No. NC_000015.9, NT_077661.3 UniGene ID Hs.459153 Ensembl Gene ID ENSG00000169594 Entrez Gene ID 646 Assay Information Unique Assay ID qHsaCID0017223 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000345382, ENST00000541809 Amplicon Context Sequence CCATAGAGCATGAGGCTGCTAATATCAAACACTACATTGGACTGGACAATCTCCA CCTGGCTTGTTGGATACATGGGGGGGATCCTTAGCTTACTTAGAGCGTGGGCCA CCCATCCATGCTTGCATTGGTCACACTGACGGTGGTTTATTTTCCCGGGTTTGAA ACTTTGG Amplicon Length (bp) 141 Chromosome Location 15:83935724-83936955 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 100 R2 0.9997 cDNA Cq 26.81 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 84.5 gDNA Cq 39.42 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report BNC1, Human Amplification -

The DNA Methylation Landscape of Glioblastoma Disease Progression Shows Extensive Heterogeneity in Time and Space
bioRxiv preprint doi: https://doi.org/10.1101/173864; this version posted August 9, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space Johanna Klughammer1*, Barbara Kiesel2,3*, Thomas Roetzer3,4, Nikolaus Fortelny1, Amelie Kuchler1, Nathan C. Sheffield5, Paul Datlinger1, Nadine Peter3,4, Karl-Heinz Nenning6, Julia Furtner3,7, Martha Nowosielski8,9, Marco Augustin10, Mario Mischkulnig2,3, Thomas Ströbel3,4, Patrizia Moser11, Christian F. Freyschlag12, Jo- hannes Kerschbaumer12, Claudius Thomé12, Astrid E. Grams13, Günther Stockhammer8, Melitta Kitzwoegerer14, Stefan Oberndorfer15, Franz Marhold16, Serge Weis17, Johannes Trenkler18, Johanna Buchroithner19, Josef Pichler20, Johannes Haybaeck21,22, Stefanie Krassnig21, Kariem Madhy Ali23, Gord von Campe23, Franz Payer24, Camillo Sherif25, Julius Preiser26, Thomas Hauser27, Peter A. Winkler27, Waltraud Kleindienst28, Franz Würtz29, Tanisa Brandner-Kokalj29, Martin Stultschnig30, Stefan Schweiger31, Karin Dieckmann3,32, Matthias Preusser3,33, Georg Langs6, Bernhard Baumann10, Engelbert Knosp2,3, Georg Widhalm2,3, Christine Marosi3,33, Johannes A. Hainfellner3,4, Adelheid Woehrer3,4#§, Christoph Bock1,34,35# 1 CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna, Austria. 2 Department of Neurosurgery, Medical University of Vienna, Vienna, Austria. 3 Comprehensive Cancer Center, Central Nervous System Tumor Unit, Medical University of Vienna, Austria. 4 Institute of Neurology, Medical University of Vienna, Vienna, Austria. 5 Center for Public Health Genomics, University of Virginia, Charlottesville VA, USA. 6 Department of Biomedical Imaging and Image-guided Therapy, Computational Imaging Research Lab, Medical University of Vi- enna, Vienna, Austria. -

Identification of Novel DNA-Methylated Genes
Prostate Cancer and Prostatic Disease (2013) 16, 292–300 & 2013 Macmillan Publishers Limited All rights reserved 1365-7852/13 www.nature.com/pcan ORIGINAL ARTICLE Identification of novel DNA-methylated genes that correlate with human prostate cancer and high-grade prostatic intraepithelial neoplasia JM Devaney1, S Wang2,3, S Funda1, J Long2, DJ Taghipour2, R Tbaishat3, P Furbert-Harris2,4, M Ittmann5 and B Kwabi-Addo2,3 BACKGROUND: Prostate cancer (PCa) harbors a myriad of genomic and epigenetic defects. Cytosine methylation of CpG-rich promoter DNA is an important mechanism of epigenetic gene inactivation in PCa. There is considerable amount of data to suggest that DNA methylation-based biomarkers may be useful for the early detection and diagnosis of PCa. In addition, candidate gene- based studies have shown an association between specific gene methylation and alterations and clinicopathologic indicators of poor prognosis in PCa. METHODS: To more comprehensively identify DNA methylation alterations in PCa initiation and progression, we examined the methylation status of 485 577 CpG sites from regions with a broad spectrum of CpG densities, interrogating both gene-associated and non-associated regions using the recently developed Illumina 450K methylation platform. RESULTS: In all, we selected 33 promoter-associated novel CpG sites that were differentially methylated in high-grade prostatic intraepithelial neoplasia and PCa in comparison with benign prostate tissue samples (false discovery rate-adjusted P-value o0.05; b-value X0.2; fold change 41.5). Of the 33 genes, hierarchical clustering analysis demonstrated BNC1, FZD1, RPL39L, SYN2, LMX1B, CXXC5, ZNF783 and CYB5R2 as top candidate novel genes that are frequently methylated and whose methylation was associated with inactivation of gene expression in PCa cell lines. -

Rna-Sequencing Applications: Gene Expression Quantification and Methylator Phenotype Identification
The Texas Medical Center Library DigitalCommons@TMC The University of Texas MD Anderson Cancer Center UTHealth Graduate School of The University of Texas MD Anderson Cancer Biomedical Sciences Dissertations and Theses Center UTHealth Graduate School of (Open Access) Biomedical Sciences 8-2013 RNA-SEQUENCING APPLICATIONS: GENE EXPRESSION QUANTIFICATION AND METHYLATOR PHENOTYPE IDENTIFICATION Guoshuai Cai Follow this and additional works at: https://digitalcommons.library.tmc.edu/utgsbs_dissertations Part of the Bioinformatics Commons, Computational Biology Commons, and the Medicine and Health Sciences Commons Recommended Citation Cai, Guoshuai, "RNA-SEQUENCING APPLICATIONS: GENE EXPRESSION QUANTIFICATION AND METHYLATOR PHENOTYPE IDENTIFICATION" (2013). The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access). 386. https://digitalcommons.library.tmc.edu/utgsbs_dissertations/386 This Dissertation (PhD) is brought to you for free and open access by the The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences at DigitalCommons@TMC. It has been accepted for inclusion in The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access) by an authorized administrator of DigitalCommons@TMC. For more information, please contact [email protected]. RNA-SEQUENCING APPLICATIONS: GENE EXPRESSION QUANTIFICATION AND METHYLATOR PHENOTYPE IDENTIFICATION -

The GALNT9, BNC1 and CCDC8 Genes Are Frequently Epigenetically Dysregulated in Breast Tumours That Metastasise to the Brain Rajendra P
Pangeni et al. Clinical Epigenetics (2015) 7:57 DOI 10.1186/s13148-015-0089-x RESEARCH Open Access The GALNT9, BNC1 and CCDC8 genes are frequently epigenetically dysregulated in breast tumours that metastasise to the brain Rajendra P. Pangeni1, Prasanna Channathodiyil1, David S. Huen2, Lawrence W. Eagles1, Balraj K. Johal2, Dawar Pasha2, Natasa Hadjistephanou2, Oliver Nevell2, Claire L. Davies2, Ayobami I. Adewumi2, Hamida Khanom2, Ikroop S. Samra2, Vanessa C. Buzatto2, Preethi Chandrasekaran2, Thoraia Shinawi3, Timothy P. Dawson4, Katherine M. Ashton4, Charles Davis4, Andrew R. Brodbelt5, Michael D. Jenkinson5, Ivan Bièche6, Farida Latif3, John L. Darling1, Tracy J. Warr1 and Mark R. Morris1,2,3* Abstract Background: Tumour metastasis to the brain is a common and deadly development in certain cancers; 18–30 % of breast tumours metastasise to the brain. The contribution that gene silencing through epigenetic mechanisms plays in these metastatic tumours is not well understood. Results: We have carried out a bioinformatic screen of genome-wide breast tumour methylation data available at The Cancer Genome Atlas (TCGA) and a broad literature review to identify candidate genes that may contribute to breast to brain metastasis (BBM). This analysis identified 82 candidates. We investigated the methylation status of these genes using Combined Bisulfite and Restriction Analysis (CoBRA) and identified 21 genes frequently methylated in BBM. We have identified three genes, GALNT9, CCDC8 and BNC1, that were frequently methylated (55, 73 and 71 %, respectively) and silenced in BBM and infrequently methylated in primary breast tumours. CCDC8 was commonly methylated in brain metastases and their associated primary tumours whereas GALNT9 and BNC1 were methylated and silenced only in brain metastases, but not in the associated primary breast tumours from individual patients. -

Identification of Candidate Tumour Suppressor Genes Frequently
Oncogene (2010) 29, 2104–2117 & 2010 Macmillan Publishers Limited All rights reserved 0950-9232/10 $32.00 www.nature.com/onc ORIGINAL ARTICLE Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma MR Morris1,2, C Ricketts1,2, D Gentle1,2, M Abdulrahman2, N Clarke3, M Brown3, T Kishida4, M Yao4, F Latif1,2 and ER Maher1,2,5 1Cancer Research UK Renal Molecular Oncology Group, University of Birmingham, Birmingham, UK; 2Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, College of Medical and Dental Sciences, University of Birmingham, Birmingham, UK; 3Genito-Urinary Cancer Research Group, Paterson Institute for Cancer Research, University of Manchester, Manchester, UK; 4Department of Urology, Yokohama City University School of Medicine, Yokohama, Japan and 5West Midlands Region Genetics Service, Birmingham Women’s Hospital, Edgbaston, Birmingham, UK Promoter region hyermethylation and transcriptional cancer are diagnosed in the world each year (Ferlay silencing is a frequent cause of tumour suppressor gene et al., 2007). The most common form of kidney cancer in (TSG) inactivation in many types of human cancers. adults is renal cell carcinoma (RCC). A majority of the Functional epigenetic studies, in which gene expression is RCC (B75%) are classified as clear cell (conventional) induced by treatment with demethylating agents, may and the next most frequent subtype is papillary RCC identify novel genes with tumour-specific methylation. We (B15% of all cases) (Mancini et al., 2008). The used high-density gene expression microarrays in a prognosis of advanced RCC is poor, although newer functional epigenetic study of 11 renal cell carcinoma treatments, based on knowledge of the molecular (RCC) cell lines. -

Genomic Anatomy of the Tyrp1 (Brown) Deletion Complex
Genomic anatomy of the Tyrp1 (brown) deletion complex Ian M. Smyth*, Laurens Wilming†, Angela W. Lee*, Martin S. Taylor*, Phillipe Gautier*, Karen Barlow†, Justine Wallis†, Sancha Martin†, Rebecca Glithero†, Ben Phillimore†, Sarah Pelan†, Rob Andrew†, Karen Holt†, Ruth Taylor†, Stuart McLaren†, John Burton†, Jonathon Bailey†, Sarah Sims†, Jan Squares†, Bob Plumb†, Ann Joy†, Richard Gibson†, James Gilbert†, Elizabeth Hart†, Gavin Laird†, Jane Loveland†, Jonathan Mudge†, Charlie Steward†, David Swarbreck†, Jennifer Harrow†, Philip North‡, Nicholas Leaves‡, John Greystrong‡, Maria Coppola‡, Shilpa Manjunath‡, Mark Campbell‡, Mark Smith‡, Gregory Strachan‡, Calli Tofts‡, Esther Boal‡, Victoria Cobley‡, Giselle Hunter‡, Christopher Kimberley‡, Daniel Thomas‡, Lee Cave-Berry‡, Paul Weston‡, Marc R. M. Botcherby‡, Sharon White*, Ruth Edgar*, Sally H. Cross*, Marjan Irvani¶, Holger Hummerich¶, Eleanor H. Simpson*, Dabney Johnson§, Patricia R. Hunsicker§, Peter F. R. Little¶, Tim Hubbard†, R. Duncan Campbell‡, Jane Rogers†, and Ian J. Jackson*ʈ *Medical Research Council Human Genetics Unit, Edinburgh EH4 2XU, United Kingdom; †Wellcome Trust Sanger Institute, and ‡Medical Research Council Rosalind Franklin Centre for Genome Research, Hinxton CB10 1SA, United Kingdom; §Life Sciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831; and ¶Department of Biochemistry, Imperial College, London SW7 2AZ, United Kingdom Communicated by Liane B. Russell, Oak Ridge National Laboratory, Oak Ridge, TN, January 9, 2006 (received for review September 15, 2005) Chromosome deletions in the mouse have proven invaluable in the deletions also provided the means to produce physical maps of dissection of gene function. The brown deletion complex com- genetic markers. Studies of this kind have been published for prises >28 independent genome rearrangements, which have several loci, including albino (Tyr), piebald (Ednrb), pink-eyed been used to identify several functional loci on chromosome 4 dilution (p), and the brown deletion complex (2–6). -

393LN V 393P 344SQ V 393P Probe Set Entrez Gene
393LN v 393P 344SQ v 393P Entrez fold fold probe set Gene Gene Symbol Gene cluster Gene Title p-value change p-value change chemokine (C-C motif) ligand 21b /// chemokine (C-C motif) ligand 21a /// chemokine (C-C motif) ligand 21c 1419426_s_at 18829 /// Ccl21b /// Ccl2 1 - up 393 LN only (leucine) 0.0047 9.199837 0.45212 6.847887 nuclear factor of activated T-cells, cytoplasmic, calcineurin- 1447085_s_at 18018 Nfatc1 1 - up 393 LN only dependent 1 0.009048 12.065 0.13718 4.81 RIKEN cDNA 1453647_at 78668 9530059J11Rik1 - up 393 LN only 9530059J11 gene 0.002208 5.482897 0.27642 3.45171 transient receptor potential cation channel, subfamily 1457164_at 277328 Trpa1 1 - up 393 LN only A, member 1 0.000111 9.180344 0.01771 3.048114 regulating synaptic membrane 1422809_at 116838 Rims2 1 - up 393 LN only exocytosis 2 0.001891 8.560424 0.13159 2.980501 glial cell line derived neurotrophic factor family receptor alpha 1433716_x_at 14586 Gfra2 1 - up 393 LN only 2 0.006868 30.88736 0.01066 2.811211 1446936_at --- --- 1 - up 393 LN only --- 0.007695 6.373955 0.11733 2.480287 zinc finger protein 1438742_at 320683 Zfp629 1 - up 393 LN only 629 0.002644 5.231855 0.38124 2.377016 phospholipase A2, 1426019_at 18786 Plaa 1 - up 393 LN only activating protein 0.008657 6.2364 0.12336 2.262117 1445314_at 14009 Etv1 1 - up 393 LN only ets variant gene 1 0.007224 3.643646 0.36434 2.01989 ciliary rootlet coiled- 1427338_at 230872 Crocc 1 - up 393 LN only coil, rootletin 0.002482 7.783242 0.49977 1.794171 expressed sequence 1436585_at 99463 BB182297 1 - up 393 -

Genome-Wide Analysis of Promoter Methylation Associated with Gene Expression Profile in Pancreatic Adenocarcinoma
Published OnlineFirst May 24, 2011; DOI: 10.1158/1078-0432.CCR-10-3431 Clinical Cancer Human Cancer Biology Research Genome-Wide Analysis of Promoter Methylation Associated with Gene Expression Profile in Pancreatic Adenocarcinoma Audrey Vincent1, Noriyuki Omura1, Seung-Mo Hong1, Andrew Jaffe4, James Eshleman1,2, and Michael Goggins1,2,3 Abstract Purpose: The goal of this study was to comprehensively identify CpG island methylation alterations between pancreatic cancers and normal pancreata and their associated gene expression alterations. Experimental Design: We employed methylated CpG island amplification followed by CpG island microarray, a method previously validated for its accuracy and reproducibility, to analyze the methylation profile of 27,800 CpG islands covering 21 MB of the human genome in nine pairs of pancreatic cancer versus normal pancreatic epithelial tissues and in three matched pairs of pancreatic cancer versus lymphoid tissues from the same individual. Results: This analysis identified 1,658 known loci that were commonly differentially methylated in pancreatic cancer compared with normal pancreas. By integrating the pancreatic DNA methylation status with the gene expression profiles of the same samples before and after treatment with the DNA methyltransferase inhibitor 5-aza-20-deoxycytidine, and the histone deacetylase inhibitor, trichostatin A, we identified dozens of aberrantly methylated and differentially expressed genes in pancreatic cancers including a more comprehensive list of hypermethylated and silenced genes that have not been previously described as targets for aberrant methylation in cancer. Conclusion: We expected that the identification of aberrantly hypermethylated and silenced genes will have diagnostic, prognostic, and therapeutic applications. Clin Cancer Res; 17(13); 4341–54. -

Basonuclin 2 Has a Function in the Multiplication of Embryonic Craniofacial Mesenchymal Cells and Is Orthologous to Disco Proteins
Basonuclin 2 has a function in the multiplication of embryonic craniofacial mesenchymal cells and is orthologous to disco proteins Amandine Vanhoutteghema, Anna Maciejewski-Duvala, Cyril Bouchea, Brigitte Delhommea, Franc¸oise Herve´ a, Fabrice Daubigneya, Guillaume Soubigoub, Masatake Arakic, Kimi Arakic, Ken-ichi Yamamurac, and Philippe Djiana,1 aUnite´Propre de Recherche 2228 du Centre National de la Recherche Scientifique, Universite´Paris Descartes, 45 rue des Saints-Pe`res, 75006 Paris, France; bInstitut Pasteur, Baˆtiment 14, 28 rue du Dr. Roux, 75015 Paris, France; and cInstitute of Resource Development and Analysis, Kumamoto University, 2-2-1, Honjo, Kumamoto 860-0811, Japan Communicated by Howard Green, Harvard Medical School, Boston, MA, June 26, 2009 (received for review January 29, 2009) Basonuclin 2 is a recently discovered zinc finger protein of unknown The genes for bnc1 and bnc2 differ greatly in size and are located function. Its paralog, basonuclin 1, is associated with the ability of on different chromosomes, but it is clear that they have a common keratinocytes to multiply. The basonuclin zinc fingers are closely evolutionary origin. Bnc1 and bnc2 are thought to possess different related to those of the Drosophila proteins disco and discorelated, but functions, since bnc2 but not bnc1 localizes to nuclear speckles the relation between disco proteins and basonuclins has remained and therefore is likely to have a function in nuclear processing elusive because the function of the disco proteins in larval head of mRNA (14). development seems to have no relation to that of basonuclin 1 and The extreme evolutionary stability of the bnc2 sequence suggests because the amino acid sequence of disco, apart from the zinc fingers, that the protein possesses an important function (2). -

The Cancer Genome Atlas Dataset-Based Analysis of Aberrantly Expressed Genes by Geneanalytics in Thymoma Associated Myasthenia Gravis: Focusing on T Cells
2323 Original Article The Cancer Genome Atlas dataset-based analysis of aberrantly expressed genes by GeneAnalytics in thymoma associated myasthenia gravis: focusing on T cells Jianying Xi1#, Liang Wang1#, Chong Yan1, Jie Song1, Yang Song2, Ji Chen2, Yongjun Zhu2, Zhiming Chen2, Chun Jin3, Jianyong Ding3, Chongbo Zhao1,4 1Department of Neurology, 2Department of Thoracic Surgery, Huashan Hospital, Fudan University, Shanghai 200040, China; 3Department of Thoracic Surgery, Zhongshan Hospital, Fudan University, Shanghai 200030, China; 4Department of Neurology, Jing’an District Centre Hospital of Shanghai, Fudan University, Shanghai 200040, China Contributions: (I) Conception and design: J Xi, L Wang, C Zhao; (II) Administrative support: J Xi, L Wang, C Zhao; (III) Provision of study materials or patients: Y Song, Y Zhu, J Chen, Z Chen, C Jin, J Ding; (IV) Collection and assembly of data: C Jin, J Ding; (V) Data analysis and interpretation: C Yan, J Song; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. #These authors contributed equally to this work. Correspondence to: Chongbo Zhao. Department of Neurology, Huashan Hospital, Fudan University, Shanghai 200040, China. Email: [email protected]. Background: Myasthenia gravis (MG) is a group of autoimmune disease which could be accompanied by thymoma. Many differences have been observed between thymoma-associated MG (TAMG) and non-MG thymoma (NMG). However, the molecular difference between them remained unknown. This study aimed to explore the differentially expressed genes (DEGs) between the two categories and to elucidate the possible pathogenesis of TAMG further. Methods: DEGs were calculated using the RNA-Sequencing data from 11 TAMG and 10 NMG in The Cancer Genome Atlas (TCGA) database. -

BNC1 Regulates Human Epicardial Heterogeneity and Function
BNC1 REGULATES HUMAN EPICARDIAL HETEROGENEITY AND FUNCTION Sophie McManus St Catharine’s College Department of Clinical Medicine/ Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre Addenbrooke’s Hospital University of Cambridge This dissertation is submitted for the degree of Doctor of Philosophy October 2019 Dedicated to A. i DECLARATION This dissertation is the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. It has not been previously submitted, in part or whole, to the University of Cambridge or any other university or institution for any degree, diploma, or other qualification. In accordance with the Department of Clinical Medicine guidelines, this thesis does not exceed 60,000 words. Sophie McManus (MA, MRes) Publications Part of the work presented in this dissertation has either been submitted or published in the following: Gambardella L., McManus S.A., Moignard V., Sebukhan D., Delaune A., Andrews S., Bernard W.G., Morrison M., Riley P.R., Le Novѐre N., Sinha S. BNC1 is a master regulator of human epicardial cell heterogeneity and function. Development, accepted for publication in August 2019. (Development 2019 146: dev174441 doi: 10.1242/dev.174441 Published 13th December 2019.) ii ABSTRACT Name: Sophie McManus Thesis title: BNC1 regulates human epicardial heterogeneity and function The epicardium is a transcriptionally heterogeneous cell layer covering the heart, crucial to correct cardiovascular development. Following epithelial-to-mesenchymal transition (EMT), epicardial cells migrate into myocardium, form coronary smooth muscle cells and cardiac fibroblasts, and instruct cardiomyocytes to proliferate and mature. Adult mammalian epicardium is quiescent, but reactivates post-injury with limited effect.