New Metal Cofactors and Recent Metallocofactor Insights Hausinger 3
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -
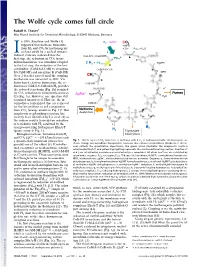
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Xuejun Yu Dissertation Final
UNIVERSITY OF CALIFORNIA RIVERSIDE Conversion of Carbon Dioxide to Formate by a Formate Dehydrogenase from Cupriavidus necator A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Bioengineering by Xuejun Yu September 2018 Dissertation Committee: Dr. Ashok Mulchandani, Co-Chairperson Dr. Xin Ge, Co-Chairperson Dr. Russ Hille Copyright by Xuejun Yu 2018 The Dissertation of Xuejun Yu is approved: Committee Co-Chairperson Committee Co-Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to express sincere appreciation to my advisor, Professor Ashok Mulchandani, for accepting me to be his student when I was suffering. Thank you very much for your delicate guidance, support and encouragement during the past years. You not only guide me on my PhD study, but also help me to be mature on my personality. From bottom of my heart, I feel very lucky to have you as professor. Also, many thanks go to my other committee members. Professor Xin Ge acted as my co-advisor and provided many valuable comments on my molecular cloning work. Professor Russ Hille has also provided insights, encouragements and advices as a member of my committee members. I have learned so many important insights from our meetings and discussions and thank you for bringing me into the molybdenum/tungsten enzyme conference. I am also thankful to the past and current group members, colleagues and friends - Dimitri Niks, Pankaj Ramnani, Feng Tan, Rabeay Hassan, Trupti Terse, Thien-Toan Tran, Claudia Chaves, Jia-wei Tay, Hui Wang, Pham Tung, Hilda Chan, Yingning Gao and Tynan Young. -

Methylotrophic Methanogenesis in Hydraulically Fractured Shales
Methylotrophic Methanogenesis in Hydraulically Fractured Shales Master’s Thesis Presented in Partial Fulfillment of the Requirements for the Degree Master of Science in the Graduate School of The Ohio State University By: Daniel Nimmer Marcus, B.S. Graduate Program in Microbiology The Ohio State University 2016 Thesis Committee: Kelly Wrighton PhD, Advisor Joseph Krzycki PhD Charles Daniels PhD Matthew Sullivan PhD Copyright by Daniel Nimmer Marcus 2016 ABSTRACT Over the last decade shale gas obtained from hydraulic fracturing of deep shale formations has become a sizeable component of the US energy portfolio. There is a growing body of evidence indicating that methanogenic archaea are both present and active in hydraulically fractured shales. However, little is known about the genomic architecture of shale derived methanogens. Here we leveraged natural gas extraction activities in the Appalachian region to gain access to fluid samples from two geographically and geologically distinct shale formations. Samples were collected over a time series from both shales for a period of greater than eleven months. Using assembly based metagenomics, two methanogen genomes from the genus Methanohalophilus were recovered and estimated to be near complete (97.1 and 100%) by 104 archaeal single copy genes. Additionally, a Methanohalophilus isolate was obtained which yielded a genome estimated to be 100% complete by the same metric. Based on metabolic reconstruction, it is inferred that these organisms utilize C-1 methyl substrates for methanogenesis. The ability to utilize monomethylamine, dimethylamine and methanol was experimentally confirmed with the Methanohalophilus isolate. In situ concentrations of C-1 methyl substrates, osmoprotectants, and Cl- were measured in parallel with estimates of community membership. -

Selenoproteins and Selenoproteomes
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Vadim Gladyshev Publications Biochemistry, Department of June 2006 Selenoproteins and selenoproteomes Vadim N. Gladyshev University of Nebraska-Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/biochemgladyshev Part of the Biochemistry, Biophysics, and Structural Biology Commons Gladyshev, Vadim N., "Selenoproteins and selenoproteomes" (2006). Vadim Gladyshev Publications. 36. https://digitalcommons.unl.edu/biochemgladyshev/36 This Article is brought to you for free and open access by the Biochemistry, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Vadim Gladyshev Publications by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Published in Selenium: Its molecular biology and role in human health, Second Edition, edited by Dolph L. Hatfi eld, Marla J. Berry, and Vadim N. Gladyshev. Springer Science+Business Me- dia LLC, 2006. Pages 99–114. Copyright © 2006 Springer Science+Business Media LLC. Used by permission. C HAPTER 9 Selenoproteins and selenoproteomes Vadim N. Gladyshev Department of Biochemistry, University of Nebraska–Lincoln Summary: In the past several years, progress in genome sequencing and development of specialized bioinformatics tools allowed effi cient identifi cation of selenocysteine-con- taining proteins encoded in completely sequenced genomes. Information is currently available on selenoproteomes from a variety of organisms, including humans, which contain 25 known selenoprotein genes. This review provides basic information about mammalian selenoproteins and other known selenoprotein families. Analysis of full sets of selenoproteins in organisms provides exciting avenues for examining selenoprotein evolution and dependence of organisms on the trace element selenium and allows link- ing selenoproteins with specifi c biological and biomedical effects of dietary selenium. -

An Unconventional Pathway for Reduction of CO2 to Methane in CO-Grown Methanosarcina Acetivorans Revealed by Proteomics
An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics Daniel J. Lessner*, Lingyun Li†, Qingbo Li*‡, Tomas Rejtar†, Victor P. Andreev†, Matthew Reichlen*, Kevin Hill*, James J. Moran§, Barry L. Karger†¶, and James G. Ferry*¶ *Department of Biochemistry and Molecular Biology and Center for Microbial Structural Biology, Pennsylvania State University, 205 South Frear Laboratory, University Park, PA 16802; †Barnett Institute and Department of Chemistry, Northeastern University, Boston, MA 02115; and §Department of Geosciences and Penn State Astrobiology Research Center, Pennsylvania State University, 220 Deike Building, University Park, PA 16802 Communicated by Lonnie O. Ingram, University of Florida, Gainesville, FL, October 5, 2006 (received for review June 27, 2006) Methanosarcina acetivorans produces acetate, formate, and methane synthesis during growth on CO (16, 22, 23). M. acetivorans was when cultured with CO as the growth substrate [Rother M, Metcalf isolated from marine sediments where giant kelp is decomposed to WW (2004) Proc Natl Acad Sci USA 101:16929–16934], which suggests methane (24). The flotation bladders of kelp contain CO that is a novel features of CO metabolism. Here we present a genome-wide presumed substrate for M. acetivorans in nature. M. acetivorans proteomic approach to identify and quantify proteins differentially produces acetate and formate in addition to methane during abundant in response to growth on CO versus methanol or acetate. CO-dependent growth (17), the first report of any product in The results indicate that oxidation of CO to CO2 supplies electrons for addition to methane and CO2 during growth of methane-producing reduction of CO2 to a methyl group by steps and enzymes of the species. -

Biochemical Pathways – Legends General Remarks for Part 1 and 2
Biochemical Pathways – Legends Copyright Boehringer Mannheim GmbH – Biochemica Courtesy of Spectrum Akademischer Verlag, GmbH General Remarks for Part 1 and 2 1) Enzymes are shown in blue The "Recommended Names" in the "Enzyme Nomenclature 1984" (Academic Press, Orlando etc.) and its supplements (Eur. J. Biochem. 157 ,1 (1986) and 179 , 489 (1989)) are used. Enzymes not listed there are printed in parentheses. 2) Coenzymes are shown in red, other reaction participants in black. 3) Color of the arrows shows type of organism in which the reaction was observed or is likely to occur: general biochemical pathways, animals, higher plants, unicellular organisms Dashed arrows are used for catabolic pathways, continuous lines for anabolic and bidirectional (amphibolic) pathways. Points on both ends of arrows indicate reversibility. An orange arrow alongside of it shows the preferred direction. 4) Regulatory effects are shown in orange. Metal ions and similar activators are located next to the reaction arrow. Effectors with a "fast" regulation of the flow (e. g. by allosteric mechanisms) have a continuous orange arrow coming from the side, those with a "slow" regulation (e. g. by induction or repression of the enzyme synthesis) a dashed orange arrow. ⊕ = increase, = decrease of enzymatic activity. Roman numerals indicate, that only one of multiple enzymes is regulated this way. Regulators shown in parentheses are effective in only one group or species of organisms. 5) Compounds enclosed in black lined, sharp edged boxes occur in several places in the chart. Coenzymes or compounds with very simple structures are not in boxes. 6) Boxes with rounded corners are sometimes used in part 2 to indicate enzymes (inactive in black, active in blue). -

Modern Metabolism As a Palimpsest of the RNA World STEVEN A
Proc. Nati. Acad. Sci. USA Vol. 86, pp. 7054-7058, September 1989 Evolution Modern metabolism as a palimpsest of the RNA world STEVEN A. BENNER*, ANDREW D. ELLINGTONt, AND ANDREAS TAUER *Laboratory for Organic Chemistry, Eidgen6ssische Technische Hochschule, CH-8092 Zurich, Switzerland; and tDepartment of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114 Communicated by F. H. Westheimer, May 15, 1989 ABSTRACT An approach is developed for constructing origin of translation, and other events that occurred in the models of ancient organisms using data from metabolic path- RNA world. ways, genetic organization, chemical structure, and enzymatic If several descendants of an ancient organism can be reaction mechanisms found in contemporary organisms. This inspected, a rule of "parsimony" can be used to model the approach is illustrated by a partial reconstruction of a model biochemistry ofthe ancestral organism by extrapolation from for the "breakthrough organism," the last organism to use the biochemistry of the descendant organisms. The most RNA as the sole genetically encoded biological catalyst. As parsimonious model is one that explains the diversity in the reconstructed here, this organism had a complex metabolism modem descendants by a minimum number of independent that included dehydrogenations, transmethylations, carbon- evolutionary events. For the progenote, three independent carbon bond-forming reactions, and an energy metabolism lineages of descendants are known (archaebacteria, eubac- based on phosphate esters. Furthermore, the breakthrough teria, and eukaryotes). Thus, a biochemical trait present in all organism probably used DNA to store genetic information, three can be assigned to the progenote. The assignment is biosynthesized porphyrins, and used terpenes as its major lipid strongest when (i) the trait is found in several representative component. -
Thioredoxin Targets Fundamental Processes in a Methane-Producing Archaeon, Methanocaldococcus Jannaschii
Thioredoxin targets fundamental processes in a methane-producing archaeon, Methanocaldococcus jannaschii Dwi Susantia,b,c, Joshua H. Wongd, William H. Vensele, Usha Loganathana,c,f, Rebecca DeSantisg,1, Ruth A. Schmitzg, Monica Balserah, Bob B. Buchanand,2, and Biswarup Mukhopadhyaya,c,f,2 Departments of aBiochemistry and fBiological Sciences, bGenetics, Bioinformatics and Computational Biology Graduate Program, and cVirginia Bioinformatics Institute, Virginia Tech, Blacksburg, VA 24061; dDepartment of Plant and Microbial Biology, University of California, Berkeley, CA 94720; eWestern Regional Research Center, United States Department of Agriculture, Agricultural Research Service, Albany, CA 94710; gInstitut für Allgemeine Mikrobiologie, Christian-Albrechts-Universität Kiel, 24118 Kiel, Germany; and hDepartamento de Estrés Abiótico, Instituto de Recursos Naturales y Agrobiología de Salamanca (IRNASA-CSIC), 37008 Salamanca, Spain Contributed by Bob B. Buchanan, January 7, 2014 (sent for review September 10, 2013) Thioredoxin (Trx), a small redox protein, controls multiple pro- strict anaerobes that produce methane, a prominent greenhouse cesses in eukaryotes and bacteria by changing the thiol redox gas and important fuel. We have focused on Methanocaldococcus status of selected proteins. The function of Trx in archaea is, jannaschii—a deeply rooted, hyperthermophilic methanogen living however, unexplored. To help fill this gap, we have investigated in deep-sea hydrothermal vents (10) where conditions mimic those this aspect in methanarchaea—strict anaerobes that produce meth- of early Earth. M. jannaschii produces methane exclusively from ane, a fuel and greenhouse gas. Bioinformatic analyses suggested H2 and CO2 via a process believed to represent an ancient form of that Trx is nearly universal in methanogens. Ancient methanogens respiration (11). -
Pyrroloquinoline Quinone - Wikipedia
Pyrroloquinoline quinone - Wikipedia Personal tools Not logged in Talk Contributions Create account Log in Pyrroloquinoline quinone From Wikipedia,Main page the free encyclopedia Contents PyrroloquinolineFeatured content quinoneNamespaces (PQQ), also called methoxatin, Views is a redoxCurrent cofactor events . It is found in soil and foods such as Pyrroloquinoline quinone [1] kiwifruitRandom, as well article as human Articlebreast milk. Enzymes containing Read PQQ areDonate called to quinoproteins.Talk Glucose dehydrogenase, one of Edit the quinoproteins,Wikipedia is used as a glucose sensor. PQQ stimulates View history growth Wikipediain bacteria. store [2] ContentsInteraction Variants Help More About Wikipedia [hide] Community Search 1 Historyportal 2 BiosynthesisRecent changes Identifiers 3 ControversyContact page regarding role as vitamin Search Wikipedia 4 See also CAS Number 72909-34-3 5 ToolsReferences 3D model (JSmol) Interactive image What links here ChEBI CHEBI:18315 [edit] HistoryRelated changes ChemSpider 997 Upload file KEGG C00113 It was discoveredSpecial pages by J.G. Hauge as the third redox cofactor after nicotinamidePermanent link and flavin in bacteria (although he MeSH PQQ+Cofactor [3] hypothesisedPage information that it was naphthoquinone). Anthony and PubChem CID 1024 ZatmanWikidata also found item the unknown redox cofactor in alcohol [4] UNII 47819QGH5L dehydrogenaseCite this page. In 1979, Salisbury and colleagues as well as Duine and colleagues[5] extracted this prosthetic group CompTox DTXSID3041162 Dashboard (EPA) fromPrint/export methanol dehydrogenase of methylotrophs and identified [show] its molecularCreate a structure. book Adachi and colleagues discovered that InChI PQQ was also found in Acetobacter.[6] Download as SMILES [show] PDF Properties BiosynthesisPrintable version [edit] Chemical formula C14H6N2O8 Languages A novel aspect of PQQ is its biosynthesis in bacteria from a Molar mass 330.208 g·mol−1 Deutsch [7] ribosomally translated precursor peptide, PqqA . -
6 EPR Signals in Methanogens
6 EPR Signals in Methanogens 6.1 Hydrogenothropic Pathway Fig. 1 shows the CO2-reducing pathway of methanogenesis, which uses H2 and CO2 as substrates. The reduction of CO2 to CH4 proceeds via coenzyme-bound C1-intermediates, methanofuran (MFR), tetrahydromethanopterin (H4MPT), and coenzyme M (2-mercaptoethanesulfonate, HS-CoM). In the first step CO2 is reduced to the level of formate: formylmethanofuran. From formylmethanofuran the formyl group is transferred to tetrahydromethanopterin, which serves as the carrier of the C1 unit during its reduction to methylene- and methyltetrahydromethanopterin. The methyl group is than transferred for the fourth and last reduction step in the pathway to a structurally simple substrate, coenzyme M. Methyl-coenzyme M (CH3-S-CoM) is reduced with coenzyme B (HS-CoB) to methane and CoM-S-S-CoB. For the reduction steps electrons from the oxidation of H2 are used. This input is either directly or via coenzyme F420. Hence the most methanogenic bacteria contain two distinct hydrogenases to oxidize H2: a coenzyme F420-reducing hydrogenase, and a coenzyme F420- non-reducing enzyme. Five enzymes of this pathway are metalloenzymes and will be discussed in the next section. CO2 H2 Formylmethanofuran Hydrogenase MFR Dehydrogenase CHO MFR H4MPT MFR CHO H4MPT CH H4MPT H2 CH2 H4MPT H2 CH3 H4MPT Methyl-H4MPT:coenzyme M H MPT Methyltransferase 4 CH3 S CoM HS CoM + HS CoB H Methyl-coenzyme M 2 Reductase Heterodisulfide CoM S S CoB Reductase CH4 Fig. 1: Metabolic pathway of methanogenesis from CO2 in Methanothermobacter marburgensis. CHO-MFR, N- 5 + 5 10 formylmethanofuran; CHO-H4MPT, N -formyltetrahydromethanopterin; CH≡H4MPT , N ,N - 5 10 + methenyltetrahydromethanopterin; CH2=H4MPT, N ,N -methylenetetrahydromethanopterin; CH3-H4MPT , N5-methyltetrahydromethanopterin; CH -S-CoM, methyl-coenzyme M. -

Methyl-Coenzyme M Reductase: Expressing Active Recombinant Enzyme in M
Methyl-coenzyme M reductase: Expressing active recombinant enzyme in M. maripaludis, and investigating the activation process, and the mechanism of inhibition of 3- nitrooxypropanol and Studying the effect of Ionic liquids on the mononuclear rearrangement of heterocycles using QM/MM method By Robel Zeray Ghebreab A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 3, 2019 Copyright 2019 by Robel Zeray Ghebreab Approved by Eduardus Cornel Duin, Chair, Professor of Chemistry and Biochemistry Holly Renee Ellis, Professor of Chemistry and Biochemistry Steven Omid Mansoorabadi, Associate Professor of Chemistry and Biochemistry Anne Elizabeth Vivian Gorden, Associate Professor of Chemistry and Biochemistry Yi Wang, Assistant Professor of Biosystems Engineering Abstract The study of the biosynthesis of methane is appealing from both its potential role in biofuel production and environmental perspectives. Methane is the main component of natural gas, and it is also a potent greenhouse gas as it is the second most abundant greenhouse gas after CO2. Methane is produced through geothermal processes and through the action of microorganisms known as methanogenic Archaea. Methanotrophic Archaea on the other hand are involved in the biological oxidation of methane. Together with aerobic methanotrophs, methanotrophic Archaea play an essential role in preventing the release of methane to the atmosphere which otherwise could have a serious contribution to global warming and climate change.1–3 Methyl-coenzyme M reductase (MCR) is the key enzyme in both the biological formation and the anaerobic oxidation of methane.4 MCR catalyzes the terminal step in methane formation and first step in the oxidation of methane.