Tungsten Biochemistry in Pyrococcus Furiosus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

Genomics and Energy and Environmental Science Poster 2011
Omics: Exploring the Molecular Universe within Biological Systems Instead of studying one or a few genes or proteins at a time, “omics” collectively Proteomics is the analysis of the proteome—the complete set describes the comprehensive analysis of genes, RNA transcripts, proteins, of proteins expressed by a cell or population of cells. Proteins, metabolites, and other molecules present in a biological system. Ongoing the workhorse molecules of life, catalyze biochemical reactions; advances in computing power and automated technologies for DNA sequencing provide structural support; and recognize, bind, or transport and experiments continue to improve our ability to analyze increasing numbers other molecules throughout the cell. Hundreds of dierent types of molecules and how they function as a system. Systems biology integrates the of proteins can be expressed at a time, and most are part of large data from various omic analyses using computational tools to build predictive complexes made up of models of biological systems. many proteins and other molecules. Transcriptomics is the analysis of the transcriptome—the complete set of RNA Hundreds to molecules present in a cell or population of thousands of different cells. RNA, which is much less stable than protein types exist within DNA, is constantly being synthesized and a cell. This large subunit of a ribosome contains about then broken down to facilitate rapid 3,000 RNA nucleotides changes in paerns of protein expression (gray) and 30 protein that occur as an organism dynamically chains (gold). responds to its environment. In addition to the three major classes of RNA (mRNA, tRNA, and ribosomal RNA), single- stranded RNA is very exible and can fold into complex shapes that carry out specic functions. -

Biological Diversity in the Patent System
Biological Diversity in the Patent System Paul Oldham1,2*, Stephen Hall1,3, Oscar Forero1,4 1 ESRC Centre for Economic and Social Aspects of Genomics (Cesagen), Lancaster University, Lancaster, United Kingdom, 2 Institute of Advanced Studies, United Nations University, Yokohama, Japan, 3 One World Analytics, Lancaster, United Kingdom, 4 Centre for Development, Environment and Policy, SOAS, University of London, London, United Kingdom Abstract Biological diversity in the patent system is an enduring focus of controversy but empirical analysis of the presence of biodiversity in the patent system has been limited. To address this problem we text mined 11 million patent documents for 6 million Latin species names from the Global Names Index (GNI) established by the Global Biodiversity Information Facility (GBIF) and Encyclopedia of Life (EOL). We identified 76,274 full Latin species names from 23,882 genera in 767,955 patent documents. 25,595 species appeared in the claims section of 136,880 patent documents. This reveals that human innovative activity involving biodiversity in the patent system focuses on approximately 4% of taxonomically described species and between 0.8–1% of predicted global species. In this article we identify the major features of the patent landscape for biological diversity by focusing on key areas including pharmaceuticals, neglected diseases, traditional medicines, genetic engineering, foods, biocides, marine genetic resources and Antarctica. We conclude that the narrow focus of human innovative activity and ownership of genetic resources is unlikely to be in the long term interest of humanity. We argue that a broader spectrum of biodiversity needs to be opened up to research and development based on the principles of equitable benefit-sharing, respect for the objectives of the Convention on Biological Diversity, human rights and ethics. -
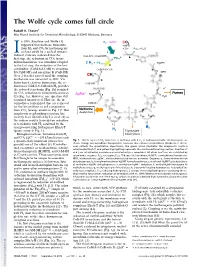
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Quantum Chemical Cluster Modeling of Enzymatic Reactions Rong-Zhen
Quantum Chemical Cluster Modeling of Enzymatic Reactions Rong-Zhen Liao 1 Rong-Zhen Liao, Stockholm, 2010 ISBN 978-91-7447-129-8 Printed in Sweden by US-AB, Stockholm 2010 Distributor: Department of Organic Chemistry, Stockholm University 2 3 4 Abstract The Quantum chemical cluster approach has been shown to be quite powerful and efficient in the modeling of enzyme active sites and reaction mechanisms. In this thesis, the reaction mechanisms of several enzymes have been investigated using the hybrid density functional B3LYP. The enzymes studied include four dinuclear zinc enzymes, namely dihydroorotase, N-acyl-homoserine lactone hydrolase, RNase Z, and human renal dipeptidase, two trinuclear zinc enzymes, namely phospholipase C and nuclease P1, two tungstoenzymes, namely formaldehyde ferredoxin oxidoreductase and acetylene hydratase, aspartate α-decarboxylase, and mycolic acid cyclopropane synthase. The potential energy profiles for various mechanistic scenarios have been calculated and analyzed. The role of the metal ions as well as important active site residues has been discussed. In the cluster approach, the effects of the parts of the enzyme that are not explicitly included in the model are taken into account using implicit solvation methods. With aspartate α-decarboxylase as an example, systematic evaluation of the solvation effects with the increase of the model size has been performed. At a model size of 150-200 atoms, the solvation effects almost vanish and the choice of the dielectric constant becomes rather insignificant. For all six zinc-dependent enzymes studied, the di-zinc bridging hydroxide has been shown to be capable of performing nucleophilic attack on the substrate. In addition, one, two, or even all three zinc ions participate in the stabilization of the negative charge in the transition states and intermediates, thereby lowering the barriers. -

And Thermo-Adaptation in Hyperthermophilic Archaea: Identification of Compatible Solutes, Accumulation Profiles, and Biosynthetic Routes in Archaeoglobus Spp
Universidade Nova de Lisboa Osmo- andInstituto thermo de Tecnologia-adaptation Química e Biológica in hyperthermophilic Archaea: Subtitle Subtitle Luís Pedro Gafeira Gonçalves Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. OH OH OH CDP c c c - CMP O O - PPi O3P P CTP O O O OH OH OH OH OH OH O- C C C O P O O P i Dissertation presented to obtain the Ph.D degree in BiochemistryO O- Instituto de Tecnologia Química e Biológica | Universidade Nova de LisboaP OH O O OH OH OH Oeiras, Luís Pedro Gafeira Gonçalves January, 2008 2008 Universidade Nova de Lisboa Instituto de Tecnologia Química e Biológica Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. This dissertation was presented to obtain a Ph. D. degree in Biochemistry at the Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa. By Luís Pedro Gafeira Gonçalves Supervised by Prof. Dr. Helena Santos Oeiras, January, 2008 Apoio financeiro da Fundação para a Ciência e Tecnologia (POCI 2010 – Formação Avançada para a Ciência – Medida IV.3) e FSE no âmbito do Quadro Comunitário de apoio, Bolsa de Doutoramento com a referência SFRH / BD / 5076 / 2001. ii ACKNOWNLEDGMENTS The work presented in this thesis, would not have been possible without the help, in terms of time and knowledge, of many people, to whom I am extremely grateful. Firstly and mostly, I need to thank my supervisor, Prof. Helena Santos, for her way of thinking science, her knowledge, her rigorous criticism, and her commitment to science. -

The Crystal Structure of Pyrococcus Furiosus Ornithine Carbamoyltransferase Reveals a Key Role for Oligomerization in Enzyme Stability at Extremely High Temperatures
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 2801–2806, March 1998 Biochemistry The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures VINCENT VILLERET*, BERNARD CLANTIN†,CATHERINE TRICOT‡,CHRISTIANNE LEGRAIN‡,MARTINE ROOVERS§¶, i VICTOR STALON†,NICOLAS GLANSDORFF‡§¶, AND JOZEF VAN BEEUMEN* *Laboratorium voor Eiwitbiochemie en Eiwitengineering, Universiteit Gent, Ledeganckstraat 35, B-9000 Gent, Belgium; and †Laboratoire de Microbiologie, Universite´Libre de Bruxelles, ‡Institut de Recherches du Centre d’Enseignement et de Recherches des Industries Alimentaires, Commission de la Communaute´ Franc¸aise de Belgique, Re´gionBruxelles Capitale, §Laboratorium voor Erfelijkheidsleer en Microbiologie, Vrije Universiteit Brussel, and ¶Vlaams Interuniversitair Instituut voor Biotechnologie, avenue E. Gryson 1, B-1070 Brussels, Belgium Edited by Max F. Perutz, Medical Research Council, Cambridge, United Kingdom, and approved January 5, 1998 (received for review September 8, 1997) ABSTRACT The Pyrococcus furiosus (PF) ornithine car- lating agent (8). The involvement of such a thermolabile bamoyltransferase (OTCase; EC 2.1.3.3) is an extremely heat- intermediate in the metabolism of extreme thermophilic mi- stable enzyme that maintains about 50% of its activity after croorganisms raises the question of which mechanisms protect heat treatment for 60 min at 100°C. To understand the it from decomposition at elevated growth temperatures. Re- molecular basis of thermostability of this enzyme, we have cent results suggest that in Thermus aquaticus and Pyrococcus determined its three-dimensional structure at a resolution of furiosus (PF), CP is protected from the bulk of the aqueous 2.7 Å and compared it with the previously reported structures phase by channeling between carbamoylphosphate synthetase of OTCases isolated from mesophilic bacteria. -

Xuejun Yu Dissertation Final
UNIVERSITY OF CALIFORNIA RIVERSIDE Conversion of Carbon Dioxide to Formate by a Formate Dehydrogenase from Cupriavidus necator A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Bioengineering by Xuejun Yu September 2018 Dissertation Committee: Dr. Ashok Mulchandani, Co-Chairperson Dr. Xin Ge, Co-Chairperson Dr. Russ Hille Copyright by Xuejun Yu 2018 The Dissertation of Xuejun Yu is approved: Committee Co-Chairperson Committee Co-Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to express sincere appreciation to my advisor, Professor Ashok Mulchandani, for accepting me to be his student when I was suffering. Thank you very much for your delicate guidance, support and encouragement during the past years. You not only guide me on my PhD study, but also help me to be mature on my personality. From bottom of my heart, I feel very lucky to have you as professor. Also, many thanks go to my other committee members. Professor Xin Ge acted as my co-advisor and provided many valuable comments on my molecular cloning work. Professor Russ Hille has also provided insights, encouragements and advices as a member of my committee members. I have learned so many important insights from our meetings and discussions and thank you for bringing me into the molybdenum/tungsten enzyme conference. I am also thankful to the past and current group members, colleagues and friends - Dimitri Niks, Pankaj Ramnani, Feng Tan, Rabeay Hassan, Trupti Terse, Thien-Toan Tran, Claudia Chaves, Jia-wei Tay, Hui Wang, Pham Tung, Hilda Chan, Yingning Gao and Tynan Young. -

Counts Metabolic Yr10.Pdf
Advanced Review Physiological, metabolic and biotechnological features of extremely thermophilic microorganisms James A. Counts,1 Benjamin M. Zeldes,1 Laura L. Lee,1 Christopher T. Straub,1 Michael W.W. Adams2 and Robert M. Kelly1* The current upper thermal limit for life as we know it is approximately 120C. Microorganisms that grow optimally at temperatures of 75C and above are usu- ally referred to as ‘extreme thermophiles’ and include both bacteria and archaea. For over a century, there has been great scientific curiosity in the basic tenets that support life in thermal biotopes on earth and potentially on other solar bodies. Extreme thermophiles can be aerobes, anaerobes, autotrophs, hetero- trophs, or chemolithotrophs, and are found in diverse environments including shallow marine fissures, deep sea hydrothermal vents, terrestrial hot springs— basically, anywhere there is hot water. Initial efforts to study extreme thermo- philes faced challenges with their isolation from difficult to access locales, pro- blems with their cultivation in laboratories, and lack of molecular tools. Fortunately, because of their relatively small genomes, many extreme thermo- philes were among the first organisms to be sequenced, thereby opening up the application of systems biology-based methods to probe their unique physiologi- cal, metabolic and biotechnological features. The bacterial genera Caldicellulosir- uptor, Thermotoga and Thermus, and the archaea belonging to the orders Thermococcales and Sulfolobales, are among the most studied extreme thermo- philes to date. The recent emergence of genetic tools for many of these organ- isms provides the opportunity to move beyond basic discovery and manipulation to biotechnologically relevant applications of metabolic engineering. -

Ts2631 Endolysin from the Extremophilic Thermus Scotoductus Bacteriophage Vb Tsc2631 As an Antimicrobial Agent Against Gram-Negative Multidrug-Resistant Bacteria
viruses Article Ts2631 Endolysin from the Extremophilic Thermus scotoductus Bacteriophage vB_Tsc2631 as an Antimicrobial Agent against Gram-Negative Multidrug-Resistant Bacteria Magdalena Plotka 1,* , Malgorzata Kapusta 2, Sebastian Dorawa 1, Anna-Karina Kaczorowska 3 and Tadeusz Kaczorowski 1,* 1 Laboratory of Extremophiles Biology, Department of Microbiology, Faculty of Biology, University of Gdansk, 80-822 Gdansk, Poland 2 Department of Plant Cytology and Embryology, Faculty of Biology, University of Gdansk, 80-308 Gdansk, Poland 3 Collection of Plasmids and Microorganisms, Faculty of Biology, University of Gdansk, 80-308 Gdansk, Poland * Correspondence: [email protected] (M.P.); [email protected] (T.K.); Tel.: +48-58-523-60-75 (M.P.); +48-58-523-60-67 (T.K.) Received: 5 June 2019; Accepted: 15 July 2019; Published: 18 July 2019 Abstract: Bacteria that thrive in extreme conditions and the bacteriophages that infect them are sources of valuable enzymes resistant to denaturation at high temperatures. Many of these heat-stable proteins are useful for biotechnological applications; nevertheless, none have been utilized as antibacterial agents. Here, we demonstrate the bactericidal potential of Ts2631 endolysin from the extremophilic bacteriophage vB_Tsc2631, which infects Thermus scotoductus, against the alarming multidrug-resistant clinical strains of Acinetobacter baumannii, Pseudomonas aeruginosa and pathogens from the Enterobacteriaceae family. A 2–3.7 log reduction in the bacterial load was observed in antibacterial tests against A. baumannii and P. aeruginosa after 1.5 h. The Ts2631 activity was further enhanced by ethylenediaminetetraacetic acid (EDTA), a metal ion chelator (4.2 log reduction in carbapenem-resistant A. baumannii) and, to a lesser extent, by malic acid and citric acid (2.9 and 3.3 log reductions, respectively). -

UNIVERSITY of EMBU EDWARD NDERITU KARANJA Phd 2020
UNIVERSITY OF EMBU EDWARD NDERITU KARANJA PhD 2020 MICROBIAL COMMUNITY DIVERSITY AND STRUCTURE WITHIN ORGANIC AND CONVENTIONAL FARMING SYSTEMS IN CENTRAL HIGHLANDS OF KENYA EDWARD NDERITU KARANJA (MSc) A THESIS SUBMITTED IN PARTIAL FULFILLMENT FOR THE DEGREE OF DOCTOR OF PHILOSOPHY IN APPLIED MICROBIOLOGY IN THE UNIVERSITY OF EMBU NOVEMBER, 2020 DECLARATION This thesis is my original work and has not been presented for a degree in any other University Signature……………………………. Date………….……….. Edward Nderitu Karanja Department of Biological Science B801/147/2015 This thesis has been submitted for examination with our approval as the University Supervisors Signature……………………………. Date………….………. Prof. Romano Mwirichia Department of Biological science University of Embu (UoEm), Kenya Signature………. …………………………. Date………….…………. Dr. Andreas Fliessbach Department of Soil Science Research Institute of Organic Agriculture - FIBL, Switzerland i DEDICATION I dedicated to my family; my wife Anne Kelly Kambura, my children; Shawn Karanja, Melissa Wangithi, Joseph Munyuithia, Shayne Koome and Ann Wanjiku, my parents; Mr. Samuel Karanja and Mrs. Agnes Wangithi, my siblings, Ruth Wairimu, Juliet Muthoni, Alex Ngochi, James Karuma and Nelly Njoki. I appreciate the support you have accorded me during my studies. Your inspiration and backing in this journey made it easier to manage all challenges encountered. ii ACKNOWLEDGEMENT I express gratitude toward Almighty God for his mercies from the beginning of this long and thought-provoking journey. This was conducted in the framework of long-term systems comparison program, with financial support from Biovision Foundation, Coop Sustainability Fund, Liechtenstein Development Service (LED) and the Swiss Agency for Development and Cooperation (SDC). I acknowledge icipe core funding for the kind contribution provided by UK-Aid from UK Government, Swedish International Development Cooperation Agency, Swiss Agency for Development and Cooperation, Federal Democratic Republic of Ethiopia and the Kenyan Government.