6 EPR Signals in Methanogens
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

One Carbon Metabolism and Its Clinical Significance
One carbon metabolism and its clinical significance Dr. Kiran Meena Department of Biochemistry Class 5 : 10-10-2019 (8:00 to 9:00 AM ) Specific Learning Objectives Describe roles of folic acid, cobalamin and S-adenosylmethionine (SAM) in transfer of one carbon units between molecules, and apply their relevance to disease states Describe synthesis of S-adenosylmethionine and its role in methylation reactions Explain how a cobalamin deficiency leads to a secondary folate deficiency Introduction Human body cannot synthesize folic acid Its also called vitamin B9 give rise to tetrahydrofolate (THF), carry one carbon groups ex. Methyl group Intestines releases mostly N5-methy-THF into blood One-carbon (1C) metabolism, mediated by folate cofactor, supports biosynthesis of purines and pyrimidines, aa homeostasis (glycine, serine and methionine) Table 14.4: DM Vasudevan’ s Textbook of Biochemistry for Medical Students 6th edition Enzyme co-factors involved in aa catabolism Involves one of three co-factors: Biotin, Tetrahydrofolate (THF) and S- adenosylmethionine (SAM) These cofactors transfer one-carbon groups in different oxidation states: 1. Biotin transfers carbon in its most oxidized state CO2, it require for catabolism and utilization of branched chain aa • Biotin responsible for carbon dioxide transfer in several carboxylase enzymes Cont-- 2. Tetrahydrofolate (THF) transfers one-carbon groups in intermediate oxidation states and as methyl groups • Tetrahydrobiopterin (BH4, THB) is a cofactor of degradation of phenylalanine • Oxidised form of THF, folate is vitamin for mammals • It converted into THF by DHF reductase 3. S-adenosylmethionine (SAM) transfers methyl groups, most reduced state of carbon THF and SAM imp in aa and nucleotide metabolism SAM used in biosynthesis of creatine, phosphatidylcholine, plasmenylcholine and epinephrine, also methylated DNA, RNA and proteins Enzymes use cobalamin as a cofactor 1. -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

A New Mode of B Binding and the Direct Participation of A
Research Article 997 A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase Naoki Shibata1, Jun Masuda1, Takamasa Tobimatsu2, Tetsuo Toraya2*, Kyoko Suto1, Yukio Morimoto1 and Noritake Yasuoka1* Background: Diol dehydratase is an enzyme that catalyzes the adenosylcobalamin Addresses: 1Department of Life Science, Himeji (coenzyme B ) dependent conversion of 1,2-diols to the corresponding Institute of Technology, 1475-2 Kanaji, Kamigori, 12 Ako-gun, Hyogo 678-1297, Japan and 2Department aldehydes. The reaction initiated by homolytic cleavage of the cobalt–carbon bond of Bioscience and Biotechnology, Faculty of α β γ of the coenzyme proceeds by a radical mechanism. The enzyme is an 2 2 2 Engineering, Okayama University, Tsushima-Naka, heterooligomer and has an absolute requirement for a potassium ion for catalytic Okayama 700-8530, Japan. activity. The crystal structure analysis of a diol dehydratase–cyanocobalamin *Corresponding authors. complex was carried out in order to help understand the mechanism of action of E-mail: [email protected] this enzyme. [email protected] Results: The three-dimensional structure of diol dehydratase in complex with Key words: B12 enzyme, diol dehydratase, radicals, cyanocobalamin was determined at 2.2 Å resolution. The enzyme exists as a reaction mechanism, TIM barrel αβγ dimer of heterotrimers ( )2. The cobalamin molecule is bound between the Received: 16 March 1999 α and β subunits in the ‘base-on’ mode, that is, 5,6-dimethylbenzimidazole of Revisions requested: 7 April 1999 the nucleotide moiety coordinates to the cobalt atom in the lower axial position. -

Mobile Loop Dynamics in Adenosyltransferase Control
Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B12 Romila Mascarenhasa,1, Markus Ruetza,1, Liam McDevitta, Markos Koutmosb,c, and Ruma Banerjeea,2 aDepartment of Biological Chemistry, University of Michigan, Ann Arbor, MI 48109-0600; bDepartment of Chemistry, University of Michigan, Ann Arbor, MI 48109-0600, and cDepartment of Biophysics, University of Michigan, Ann Arbor, MI 48109-0600 Edited by Amie K. Boal, Pennsylvania State University, State College, PA, and accepted by Editorial Board Member Stephen J. Benkovic October 20, 2020 (received for review April 16, 2020) Cobalamin is a complex organometallic cofactor that is processed Human and Methylobacterium extorquens (Me) ATR, which have and targeted via a network of chaperones to its dependent been characterized most extensively, also double as chaperones, enzymes. AdoCbl (5′-deoxyadenosylcobalamin) is synthesized transferring AdoCbl directly to MCM (Fig. 1A) rather than re- from cob(II)alamin in a reductive adenosylation reaction catalyzed leasing the high-value product into solution (12–17). Cofactor by adenosyltransferase (ATR), which also serves as an escort, de- loading onto MCM is gated by the GTPase activity of the G livering AdoCbl to methylmalonyl-CoA mutase (MCM). The mech- protein chaperone CblA (18). Despite the overall structural and anism by which ATR signals that its cofactor cargo is ready functional conservation of the cofactor loading processes, there (AdoCbl) or not [cob(II)alamin] for transfer to MCM, is not known. are important differences in regulation between the human (19) In this study, we have obtained crystallographic snapshots that – reveal ligand-induced ordering of the N terminus of Mycobacte- and the better-characterized bacterial systems (15 17). -
The Wolfe Cycle Comes Full Circle
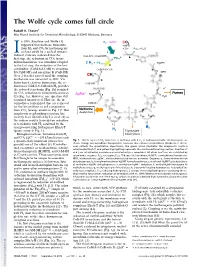
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Annotation Guidelines for Experimental Procedures
Annotation Guidelines for Experimental Procedures Developed By Mohammed Alliheedi Robert Mercer Version 1 April 14th, 2018 1- Introduction and background information What is rhetorical move? A rhetorical move can be defined as a text fragment that conveys a distinct communicative goal, in other words, a sentence that implies an author’s specific purpose to readers. What are the types of rhetorical moves? There are several types of rhetorical moves. However, we are interested in 4 rhetorical moves that are common in the method section of a scientific article that follows the Introduction Methods Results and Discussion (IMRaD) structure. 1- Description of a method: It is concerned with a sentence(s) that describes experimental events (e.g., “Beads with bound proteins were washed six times (for 10 min under rotation at 4°C) with pulldown buffer and proteins harvested in SDS-sample buffer, separated by SDS-PAGE, and analyzed by autoradiography.” (Ester & Uetz, 2008)). 2- Appeal to authority: It is concerned with a sentence(s) that discusses the use of standard methods, protocols, and procedures. There are two types of this move: - A reference to a well-established “standard” method (e.g., the use of a method like “PCR” or “electrophoresis”). - A reference to a method that was previously described in the literature (e.g., “Protein was determined using fluorescamine assay [41].” (Larsen, Frandesn and Treiman, 2001)). 3- Source of materials: It is concerned with a sentence(s) that lists the source of biological materials that are used in the experiment (e.g., “All microalgal strains used in this study are available at the Elizabeth Aidar Microalgae Culture Collection, Department of Marine Biology, Federal Fluminense University, Brazil.” (Larsen, Frandesn and Treiman, 2001)). -

Chemistry and Functions of Proteins
TVER STATE MEDICAL UNIVERSITY BIOCHEMISTRY DEPARTMENT CHEMISTRY AND FUNCTIONS OF PROTEINS ILLUSTRATED BIOCHEMISTRY Schemes, formulas, terms and algorithm of preparation The manual for making notes of lectures and preparation for classes Tver, 2018 AMINO ACIDS -amino acids -These are organic acids with at least a minimum of one of its hydrogen atoms in the carbon chains substituted by an amino group.( Show the radical, amino and carboxyl groups) NH2 R | C – H | COOH Proteinogenous and Nonproteinogenous Amino acids - Major proteinogenous (standard) amino acids. (Give the names of each amino acid) R R 1 H – 2 CH3 – 12 HO – CH2 – (CH3)2 CH – 3 CH3 – CH – (CH3)2 CH – CH2 – 4 13 | CH3 – CH2 – CH – OH 5 | CH 3 6 14 HS – CH2 – HOOC – CH2 – 15 CH3 – S – CH2 – CH2 – 7 HOOC – CH2 – CH2 – NH2 | – C – H - | COOH 8 16 NH2 – CO – CH2 – 9 NH2 – CO – CH2 – CH2 – 17 10 NH2 – (CH2)3 – CH2 – 18 NH2 – C – NH – (CH2)3 – CH2 – 11 || NH 19 20 СООН NH -Glycine -Arginine Alanine -Serine -Valine -Threonine Leucine -Cysteine Isoleucine -Methionine Aspartic acid -Phenylalanine Glutamic acid -Tyrosine Asparagine -Tryptophan Glutamine -Histidine Lysine -Proline •Rare proteinogenous (standard) amino acids. ( Derivatives of lysine, proline and tyrosine) NH2 | H2N – CH2 – CH – (CH2)2 – CH | | OH COOH •Nonproteinogenous amino acids. (Name and show them) NH2 NH2 | | H2N – (CH2)3 – CH HS – (CH2)2 – CH | | COOH COOH NH2 | NH2 – C – HN – (CH2)3 – CH || | O COOH Ornithine Homocysteine Citrulline 2 CLASSIFICATION OF PROTEINOGENOUS (STANDARD) AMINO ACIDS Amino acids are classified by: *The structure of the radical (show); - aliphatic amino acids -monoaminodicarboxylic amino acids -amides of amino acids -diaminomonocarboxylic amino acids -hydroxy amino acids -sulfur-containing amino acids -cyclic (aromatic and heterolytic) amino acids *the polarity of the radical (show); -Non-polar (hydrophobic –Ala, Val, Leu, Ile, Trp, Pro.) -Polar (hydrophilic). -

Chapter 7. "Coenzymes and Vitamins" Reading Assignment
Chapter 7. "Coenzymes and Vitamins" Reading Assignment: pp. 192-202, 207-208, 212-214 Problem Assignment: 3, 4, & 7 I. Introduction Many complex metabolic reactions cannot be carried out using only the chemical mechanisms available to the side-chains of the 20 standard amino acids. To perform these reactions, enzymes must rely on other chemical species known broadly as cofactors that bind to the active site and assist in the reaction mechanism. An enzyme lacking its cofactor is referred to as an apoenzyme whereas the enzyme with its cofactor is referred to as a holoenzyme. Cofactors are subdivided into essential ions and organic molecules known as coenzymes (Fig. 7.1). Essential ions, commonly metal ions, may participate in substrate binding or directly in the catalytic mechanism. Coenzymes typically act as group transfer agents, carrying electrons and chemical groups such as acyl groups, methyl groups, etc., depending on the coenzyme. Many of the coenzymes are derived from vitamins which are essential for metabolism, growth, and development. We will use this chapter to introduce all of the vitamins and coenzymes. In a few cases--NAD+, FAD, coenzyme A--the mechanisms of action will be covered. For the remainder of the water-soluble vitamins, discussion of function will be delayed until we encounter them in metabolism. We also will discuss the biochemistry of the fat-soluble vitamins here. II. Inorganic cation cofactors Many enzymes require metal cations for activity. Metal-activated enzymes require or are stimulated by cations such as K+, Ca2+, or Mg2+. Often the metal ion is not tightly bound and may even be carried into the active site attached to a substrate, as occurs in the case of kinases whose actual substrate is a magnesium-ATP complex. -

INFORMATION to USERS the Most Advanced Technology Has Been
INFORMATION TO USERS The most advanced technology has been used to photo graph and reproduce this manuscript from the microfilm master. UMI films the original text directly from the copy submitted. Thus, some dissertation copies are in typewriter face, while others may be from a computer printer. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyrighted material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are re produced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. Each oversize page is available as one exposure on a standard 35 mm slide or as a 17" x 23" black and white photographic print for an additional charge. Photographs included in the original manuscript have been reproduced xerographically in this copy. 35 mm slides or 6" x 9" black and white photographic prints are available for any photographs or illustrations appearing in this copy for an additional charge. Contact UMI directly to order. Accessing the World'sUMI Information since 1938 300 North Zeeb Road, Ann Arbor, Ml 48106-1346 USA Order Number 8820378 Stereochemical studies in anaerobic metabolism Zydowsky, Lynne Douthit, Ph.D. The Ohio State University, 1988 UMI 300 N. Zeeb Rd. Ann Aibor, M I 48106 PLEASE NOTE: In all cases this material has been filmed in the best possible way from the available copy. Problems encountered with this document have been identified here with a check mark V . -

Recycling of Vitamin B12 and NAD+ Within the Pdu Microcompartment of Salmonella Enterica Shouqiang Cheng Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2010 Recycling of vitamin B12 and NAD+ within the Pdu microcompartment of Salmonella enterica Shouqiang Cheng Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Biochemistry, Biophysics, and Structural Biology Commons Recommended Citation Cheng, Shouqiang, "Recycling of vitamin B12 and NAD+ within the Pdu microcompartment of Salmonella enterica" (2010). Graduate Theses and Dissertations. 11713. https://lib.dr.iastate.edu/etd/11713 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. + Recycling of vitamin B12 and NAD within the Pdu microcompartment of Salmonella enterica by Shouqiang Cheng A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Biochemistry Program of Study Committee: Thomas A. Bobik, Major Professor Alan DiSpirito Basil Nikolau Reuben Peters Gregory J. Phillips Iowa State University Ames, Iowa 2010 Copyright © Shouqiang Cheng, 2010. All rights reserved. ii Table of contents Abstract............................................................................................................................. -

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection. -

Coenzymes and Prosthetic Groups Nomenclature
Coenzymes and prosthetic groups Nomenclature • Cofactor: nonprotein component of enzymes • Cofactor - a co-catalyst required for enzyme activity • Coenzyme - a dissociable cofactor, usually organic • Prosthetic group - non-dissociable cofactor • Vitamin - a required micro-nutrient (organism cannot synthesize adequate quantities for normal health - may vary during life-cycle). – water soluble - not stored, generally no problem with overdose – lipid soluble - stored, often toxic with overdose. • Apoenzyme - enzyme lacking cofactor (inactive) • Holoenzyme - enzyme with cofactors (active) Vitamins are precursors of cofactors Why cofactors? Adenine Nucleotide Coenzymes All use the adenine nucleotide group solely for binding to the enzyme! • pyridine dinucleotides (NADH, NADPH) • flavin mono- and dinucleotides (FMN, FADH) • coenzyme A Nucleotide triphosphates • ATP hydrolysis – resonance stabilizes products – reactants cannot be resonance stabilized because of competition with adjacent bridging anhydrides – charge density greater on reactants than products Coenzyme A • Activation of acyl groups for transfer by nucleophilic attack • activation of the alpha- hydrogen of the acyl group for abstraction as a proton • Both these functions are mediated by the reactive -SH group on CoA, which forms thioesters Coenzyme A Nicotinic Acid/Nicotinamide Coenzymes • These coenzymes are two-electron carriers • They transfer hydride anion (H-) to and from substrates • Two important coenzymes in this class: • Nicotinamide adenine dinucleotide (NAD+) • Nicotinamide