Methanogenic Archaea Use a Bacteria-Like Methyltransferase System to Demethoxylate Aromatic Compounds
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

A Web Tool for Hydrogenase Classification and Analysis Dan Søndergaard1, Christian N
www.nature.com/scientificreports OPEN HydDB: A web tool for hydrogenase classification and analysis Dan Søndergaard1, Christian N. S. Pedersen1 & Chris Greening2,3 H2 metabolism is proposed to be the most ancient and diverse mechanism of energy-conservation. The Received: 24 June 2016 metalloenzymes mediating this metabolism, hydrogenases, are encoded by over 60 microbial phyla Accepted: 09 September 2016 and are present in all major ecosystems. We developed a classification system and web tool, HydDB, Published: 27 September 2016 for the structural and functional analysis of these enzymes. We show that hydrogenase function can be predicted by primary sequence alone using an expanded classification scheme (comprising 29 [NiFe], 8 [FeFe], and 1 [Fe] hydrogenase classes) that defines 11 new classes with distinct biological functions. Using this scheme, we built a web tool that rapidly and reliably classifies hydrogenase primary sequences using a combination of k-nearest neighbors’ algorithms and CDD referencing. Demonstrating its capacity, the tool reliably predicted hydrogenase content and function in 12 newly-sequenced bacteria, archaea, and eukaryotes. HydDB provides the capacity to browse the amino acid sequences of 3248 annotated hydrogenase catalytic subunits and also contains a detailed repository of physiological, biochemical, and structural information about the 38 hydrogenase classes defined here. The database and classifier are freely and publicly available at http://services.birc.au.dk/hyddb/ Microorganisms conserve energy by metabolizing H2. Oxidation of this high-energy fuel yields electrons that can be used for respiration and carbon-fixation. This diffusible gas is also produced in diverse fermentation and 1 anaerobic respiratory processes . H2 metabolism contributes to the growth and survival of microorganisms across the three domains of life, including chemotrophs and phototrophs, lithotrophs and heterotrophs, aerobes and 1,2 anaerobes, mesophiles and extremophiles alike . -

Evaluation of Total Protein Production by Soil Cyanobacteria in Culture Filtrate at Various Incubations Periods
www.ijapbc.com IJAPBC – Vol. 5(3), Jul - Sep, 2016 ISSN: 2277 - 4688 INTERNATIONAL JOURNAL OF ADVANCES IN PHARMACY, BIOLOGY AND CHEMISTRY Research Article Evaluation of Total Protein Production by soil Cyanobacteria in culture filtrate at various Incubations periods Farida P. Minocheherhomji* and Aarti Pradhan *Department of Microbiology, B. P. Baria Science Institute, Navsari, Gujarat, India - 396445. ABSTRACT Cyanobacteria, also known as blue-green algae, are microscopic organisms that obtain their energy through photosynthesis, and are found in common and naturally occurring ecosystem sites like moist soil and water bodies. Cyanobacteria are in a range of shapes and sizes and can occur as single cells while others assemble into groups as colonies or filaments. Blue green algae produces many metabolites including amino acids, proteins, vitamins and plant growth regulators like auxins, gibberellins and abscisic acids. The present study has been undertaken to estimate the total protein in their culture filtrate during different incubation times using BG-11 broth and Pringsheim’s broth. Protein was estimated from culture filtrate by standard protocols. The present study revealed that the amount of biomass, protein and IAA by different Cyanobacterial species were increased with the corresponding incubation time, and showed maximum concentration of 24 μg/ml, 14 μg/ml and 32.5 μg/ml in Pringsheim’s broth after 30 days respectively. Key Words: Cyanobacteria, Nitrogen fixation and Protein production INTRODUCTION Cyanobacteria belong to the group of organisms repertoire of metabolic activities. They proliferate in called prokaryotes, which also includes bacteria, and diverse types of ecosystems - ranging from the cold can be regarded as simple in terms of their cell Tundra to the hot deserts, from surface waters of structure. -
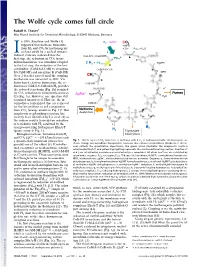
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Archaeoglobus Profundus Type Strain (AV18T)
Standards in Genomic Sciences (2010) 2:327-346 DOI:10.4056/sigs.942153 Complete genome sequence of Archaeoglobus profundus type strain (AV18T) Mathias von Jan1, Alla Lapidus2, Tijana Glavina Del Rio2, Alex Copeland2, Hope Tice2, Jan-Fang Cheng2, Susan Lucas2, Feng Chen2, Matt Nolan2, Lynne Goodwin2,3, Cliff Han2,3, Sam Pitluck2, Konstantinos Liolios2, Natalia Ivanova2, Konstantinos Mavromatis2, Galina Ovchinnikova2, Olga Chertkov2, Amrita Pati2, Amy Chen4, Krishna Palaniappan4, Miriam Land2,5, Loren Hauser2,5, Yun-Juan Chang2,5, Cynthia D. Jeffries2,5, Elizabeth Saunders2, Thomas Brettin2,3, John C. Detter2,3, Patrick Chain2,4, Konrad Eichinger6, Harald Huber6, Ste- fan Spring1, Manfred Rohde7, Markus Göker1, Reinhard Wirth6, Tanja Woyke2, Jim Bristow2, Jonathan A. Eisen2,8, Victor Markowitz4, Philip Hugenholtz2, Nikos C Kyrpides2, and Hans-Peter Klenk1* 1 DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany 2 DOE Joint Genome Institute, Walnut Creek, California, USA 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA 4 Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, California, USA 5 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA 6 University of Regensburg, Microbiology – Archaeenzentrum, Regensburg, Germany 7 HZI – Helmholtz Centre for Infection Research, Braunschweig, Germany 8 University of California Davis Genome Center, Davis, California, USA *Corresponding author: Hans-Peter Klenk Keywords: hyperthermophilic, marine, strictly anaerobic, sulfate respiration, hydrogen utili- zation, hydrothermal systems, Archaeoglobaceae, GEBA Archaeoglobus profundus (Burggraf et al. 1990) is a hyperthermophilic archaeon in the eu- ryarchaeal class Archaeoglobi, which is currently represented by the single family Archaeog- lobaceae, containing six validly named species and two strains ascribed to the genus 'Geoglobus' which is taxonomically challenged as the corresponding type species has no va- lidly published name. -

Assessment of the Carbon Monoxide Metabolism of the Hyperthermophilic Sulfate-Reducing Archaeon Archaeoglobus Fulgidus VC-16 by Comparative Transcriptome Analyses
Hindawi Publishing Corporation Archaea Volume 2015, Article ID 235384, 12 pages http://dx.doi.org/10.1155/2015/235384 Research Article Assessment of the Carbon Monoxide Metabolism of the Hyperthermophilic Sulfate-Reducing Archaeon Archaeoglobus fulgidus VC-16 by Comparative Transcriptome Analyses William P. Hocking, Irene Roalkvam, Carina Magnussen, Runar Stokke, and Ida H. Steen Department of Biology, Centre for Geobiology, University of Bergen, 5020 Bergen, Norway Correspondence should be addressed to Ida H. Steen; [email protected] Received 10 April 2015; Revised 9 June 2015; Accepted 14 June 2015 Academic Editor: Uwe Deppenmeier Copyright © 2015 William P. Hocking et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The hyperthermophilic, sulfate-reducing archaeon, Archaeoglobus fulgidus, utilizes CO as an energy source and it is resistant to the toxic effects of high CO concentrations. Herein, transcription profiles were obtained from A. fulgidus during growth with CO and sulfate or thiosulfate, or without an electron acceptor. This provided a basis for a model of the CO metabolism of A. fulgidus. The model suggests proton translocation by “Mitchell-type” loops facilitated by Fqo catalyzingred aFd :menaquinone oxidoreductase reaction, as the major mode of energy conservation, rather than formate or H2 cycling during respiratory growth. The bifunctional CODH (cdhAB-2) is predicted to play an ubiquitous role in the metabolism of CO, and a novel nitrate reductase- associated respiratory complex was induced specifically in the presence of sulfate. A potential role of this complex in relation to Fdred and APS reduction is discussed. -

Structure and Function of the Archaeal Response Regulator Chey
Structure and function of the archaeal response PNAS PLUS regulator CheY Tessa E. F. Quaxa, Florian Altegoerb, Fernando Rossia, Zhengqun Lia, Marta Rodriguez-Francoc, Florian Krausd, Gert Bangeb,1, and Sonja-Verena Albersa,1 aMolecular Biology of Archaea, Faculty of Biology, University of Freiburg, 79104 Freiburg, Germany; bLandes-Offensive zur Entwicklung Wissenschaftlich- ökonomischer Exzellenz Center for Synthetic Microbiology & Faculty of Chemistry, Philipps-University-Marburg, 35043 Marburg, Germany; cCell Biology, Faculty of Biology, University of Freiburg, 79104 Freiburg, Germany; and dFaculty of Chemistry, Philipps-University-Marburg, 35043 Marburg, Germany Edited by Norman R. Pace, University of Colorado at Boulder, Boulder, CO, and approved December 13, 2017 (received for review October 2, 2017) Motility is a central feature of many microorganisms and provides display different swimming mechanisms. Counterclockwise ro- an efficient strategy to respond to environmental changes. Bacteria tation results in smooth swimming in well-characterized peritri- and archaea have developed fundamentally different rotary motors chously flagellated bacteria such as Escherichia coli, whereas enabling their motility, termed flagellum and archaellum, respec- rotation in the opposite direction results in tumbling. In contrast, tively. Bacterial motility along chemical gradients, called chemo- in other bacteria (i.e., Vibrio alginolyticus) and haloarchaea, taxis, critically relies on the response regulator CheY, which, when clockwise rotation results -

Annotation Guidelines for Experimental Procedures
Annotation Guidelines for Experimental Procedures Developed By Mohammed Alliheedi Robert Mercer Version 1 April 14th, 2018 1- Introduction and background information What is rhetorical move? A rhetorical move can be defined as a text fragment that conveys a distinct communicative goal, in other words, a sentence that implies an author’s specific purpose to readers. What are the types of rhetorical moves? There are several types of rhetorical moves. However, we are interested in 4 rhetorical moves that are common in the method section of a scientific article that follows the Introduction Methods Results and Discussion (IMRaD) structure. 1- Description of a method: It is concerned with a sentence(s) that describes experimental events (e.g., “Beads with bound proteins were washed six times (for 10 min under rotation at 4°C) with pulldown buffer and proteins harvested in SDS-sample buffer, separated by SDS-PAGE, and analyzed by autoradiography.” (Ester & Uetz, 2008)). 2- Appeal to authority: It is concerned with a sentence(s) that discusses the use of standard methods, protocols, and procedures. There are two types of this move: - A reference to a well-established “standard” method (e.g., the use of a method like “PCR” or “electrophoresis”). - A reference to a method that was previously described in the literature (e.g., “Protein was determined using fluorescamine assay [41].” (Larsen, Frandesn and Treiman, 2001)). 3- Source of materials: It is concerned with a sentence(s) that lists the source of biological materials that are used in the experiment (e.g., “All microalgal strains used in this study are available at the Elizabeth Aidar Microalgae Culture Collection, Department of Marine Biology, Federal Fluminense University, Brazil.” (Larsen, Frandesn and Treiman, 2001)). -

Untargetted Metabolomic Exploration of the Mycobacterium Tuberculosis Stress Response to Cinnamon Essential Oil
biomolecules Article Untargetted Metabolomic Exploration of the Mycobacterium tuberculosis Stress Response to Cinnamon Essential Oil Elwira Sieniawska 1,* , Rafał Sawicki 2 , Joanna Golus 2 and Milen I. Georgiev 3,4 1 Chair and Department of Pharmacognosy, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland 2 Chair and Department of Biochemistry and Biotechnology, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland; [email protected] (R.S.); [email protected] (J.G.) 3 Group of Plant Cell Biotechnology and Metabolomics, The Stephan Angeloff Institute of Microbiology, Bulgarian Academy of Sciences, 139 Ruski Blvd., 4000 Plovdiv, Bulgaria; [email protected] 4 Center of Plant Systems Biology and Biotechnology, 4000 Plovdiv, Bulgaria * Correspondence: [email protected] Received: 6 January 2020; Accepted: 24 February 2020; Published: 26 February 2020 Abstract: The antimycobacterial activity of cinnamaldehyde has already been proven for laboratory strains and for clinical isolates. What is more, cinnamaldehyde was shown to threaten the mycobacterial plasma membrane integrity and to activate the stress response system. Following promising applications of metabolomics in drug discovery and development we aimed to explore the mycobacteria response to cinnamaldehyde within cinnamon essential oil treatment by untargeted liquid chromatography–mass spectrometry. The use of predictive metabolite pathway analysis and description of produced lipids enabled the evaluation of the stress symptoms shown by bacteria. This study suggests that bacteria exposed to cinnamaldehyde could reorganize their outer membrane as a physical barrier against stress factors. They probably lowered cell wall permeability and inner membrane fluidity, and possibly redirected carbon flow to store energy in triacylglycerols. Being a reactive compound, cinnamaldehyde may also contribute to disturbances in bacteria redox homeostasis and detoxification mechanisms. -

Xuejun Yu Dissertation Final
UNIVERSITY OF CALIFORNIA RIVERSIDE Conversion of Carbon Dioxide to Formate by a Formate Dehydrogenase from Cupriavidus necator A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Bioengineering by Xuejun Yu September 2018 Dissertation Committee: Dr. Ashok Mulchandani, Co-Chairperson Dr. Xin Ge, Co-Chairperson Dr. Russ Hille Copyright by Xuejun Yu 2018 The Dissertation of Xuejun Yu is approved: Committee Co-Chairperson Committee Co-Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to express sincere appreciation to my advisor, Professor Ashok Mulchandani, for accepting me to be his student when I was suffering. Thank you very much for your delicate guidance, support and encouragement during the past years. You not only guide me on my PhD study, but also help me to be mature on my personality. From bottom of my heart, I feel very lucky to have you as professor. Also, many thanks go to my other committee members. Professor Xin Ge acted as my co-advisor and provided many valuable comments on my molecular cloning work. Professor Russ Hille has also provided insights, encouragements and advices as a member of my committee members. I have learned so many important insights from our meetings and discussions and thank you for bringing me into the molybdenum/tungsten enzyme conference. I am also thankful to the past and current group members, colleagues and friends - Dimitri Niks, Pankaj Ramnani, Feng Tan, Rabeay Hassan, Trupti Terse, Thien-Toan Tran, Claudia Chaves, Jia-wei Tay, Hui Wang, Pham Tung, Hilda Chan, Yingning Gao and Tynan Young. -

Depth-Stratified Functional and Taxonomic Niche Specialization in the ‘Core’ and ‘Flexible’ Pacific Ocean Virome
The ISME Journal (2015) 9, 472–484 & 2015 International Society for Microbial Ecology All rights reserved 1751-7362/15 www.nature.com/ismej ORIGINAL ARTICLE Depth-stratified functional and taxonomic niche specialization in the ‘core’ and ‘flexible’ Pacific Ocean Virome Bonnie L Hurwitz1, Jennifer R Brum and Matthew B Sullivan Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ, USA Microbes drive myriad ecosystem processes, and their viruses modulate microbial-driven processes through mortality, horizontal gene transfer, and metabolic reprogramming by viral-encoded auxiliary metabolic genes (AMGs). However, our knowledge of viral roles in the oceans is primarily limited to surface waters. Here we assess the depth distribution of protein clusters (PCs) in the first large- scale quantitative viral metagenomic data set that spans much of the pelagic depth continuum (the Pacific Ocean Virome; POV). This established ‘core’ (180 PCs; one-third new to science) and ‘flexible’ (423K PCs) community gene sets, including niche-defining genes in the latter (385 and 170 PCs are exclusive and core to the photic and aphotic zones, respectively). Taxonomic annotation suggested that tailed phages are ubiquitous, but not abundant (o5% of PCs) and revealed depth- related taxonomic patterns. Functional annotation, coupled with extensive analyses to document non-viral DNA contamination, uncovered 32 new AMGs (9 core, 20 photic and 3 aphotic) that introduce ways in which viruses manipulate infected host metabolism, and parallel depth-stratified host adaptations (for example, photic zone genes for iron–sulphur cluster modulation for phage production, and aphotic zone genes for high-pressure deep-sea survival). Finally, significant vertical flux of photic zone viruses to the deep sea was detected, which is critical for interpreting depth- related patterns in nature. -

Cell Structure and Function in the Bacteria and Archaea
4 Chapter Preview and Key Concepts 4.1 1.1 DiversityThe Beginnings among theof Microbiology Bacteria and Archaea 1.1. •The BacteriaThe are discovery classified of microorganismsinto several Cell Structure wasmajor dependent phyla. on observations made with 2. theThe microscope Archaea are currently classified into two 2. •major phyla.The emergence of experimental 4.2 Cellscience Shapes provided and Arrangements a means to test long held and Function beliefs and resolve controversies 3. Many bacterial cells have a rod, spherical, or 3. MicroInquiryspiral shape and1: Experimentation are organized into and a specific Scientificellular c arrangement. Inquiry in the Bacteria 4.31.2 AnMicroorganisms Overview to Bacterialand Disease and Transmission Archaeal 4.Cell • StructureEarly epidemiology studies suggested how diseases could be spread and 4. Bacterial and archaeal cells are organized at be controlled the cellular and molecular levels. 5. • Resistance to a disease can come and Archaea 4.4 External Cell Structures from exposure to and recovery from a mild 5.form Pili allowof (or cells a very to attach similar) to surfacesdisease or other cells. 1.3 The Classical Golden Age of Microbiology 6. Flagella provide motility. Our planet has always been in the “Age of Bacteria,” ever since the first 6. (1854-1914) 7. A glycocalyx protects against desiccation, fossils—bacteria of course—were entombed in rocks more than 3 billion 7. • The germ theory was based on the attaches cells to surfaces, and helps observations that different microorganisms years ago. On any possible, reasonable criterion, bacteria are—and always pathogens evade the immune system. have been—the dominant forms of life on Earth.