Methane Utilization in Methylomicrobium Alcaliphilum 20ZR: a Systems Approach Received: 8 September 2017 Ilya R
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

Hydrogen Isotope Fractionation in Lipids of the Methane-Oxidizing Bacterium Methylococcus Capsulatus
Geochimica et Cosmochimica Acta, Vol. 66, No. 22, pp. 3955–3969, 2002 Copyright © 2002 Elsevier Science Ltd Pergamon Printed in the USA. All rights reserved 0016-7037/02 $22.00 ϩ .00 PII S0016-7037(02)00981-X Hydrogen isotope fractionation in lipids of the methane-oxidizing bacterium Methylococcus capsulatus 1, 2 3 1 ALEX L. SESSIONS, *LINDA L. JAHNKE, ARNDT SCHIMMELMANN, and JOHN M. HAYES 1Department of Geology and Geophysics, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA 2Exobiology Branch, NASA-Ames Research Center, Moffett Field, CA 94035, USA 3Biogeochemical Laboratories, Department of Geological Sciences, Indiana University, Bloomington, IN 47405, USA (Received December 10, 2001; accepted in revised form June 7, 2002) Abstract—Hydrogen isotopic compositions of individual lipids from Methylococcus capsulatus, an aerobic, methane-oxidizing bacterium, were analyzed by hydrogen isotope-ratio-monitoring gas chromatography–mass spectrometry (GC-MS). The purposes of the study were to measure isotopic fractionation factors between methane, water, and lipids and to examine the biochemical processes that determine the hydrogen isotopic composition of lipids. M. capsulatus was grown in six replicate cultures in which the ␦D values of methane and water were varied independently. Measurement of concomitant changes in ␦D values of lipids allowed estimation of the proportion of hydrogen derived from each source and the isotopic fractionation associated with the utilization of each source. All lipids examined, including fatty acids, sterols, and hopanols, derived 31.4 Ϯ 1.7% of their hydrogen from methane. This was apparently true whether the cultures were harvested during exponential or stationary phase. Examination of the relevant biochemical pathways indicates that no hydrogen is transferred directly (with C-H bonds intact) from methane to lipids. -

Reduced Net Methane Emissions Due to Microbial Methane Oxidation in a Warmer Arctic
LETTERS https://doi.org/10.1038/s41558-020-0734-z Reduced net methane emissions due to microbial methane oxidation in a warmer Arctic Youmi Oh 1, Qianlai Zhuang 1,2,3 ✉ , Licheng Liu1, Lisa R. Welp 1,2, Maggie C. Y. Lau4,9, Tullis C. Onstott4, David Medvigy 5, Lori Bruhwiler6, Edward J. Dlugokencky6, Gustaf Hugelius 7, Ludovica D’Imperio8 and Bo Elberling 8 Methane emissions from organic-rich soils in the Arctic have bacteria (methanotrophs) and the remainder is mostly emitted into been extensively studied due to their potential to increase the atmosphere (Fig. 1a). The methanotrophs in these wet organic the atmospheric methane burden as permafrost thaws1–3. soils may be low-affinity methanotrophs (LAMs) that require However, this methane source might have been overestimated >600 ppm of methane (by moles) for their growth and mainte- without considering high-affinity methanotrophs (HAMs; nance23. But in dry mineral soils, the dominant methanotrophs are methane-oxidizing bacteria) recently identified in Arctic min- high-affinity methanotrophs (HAMs), which can survive and grow 4–7 eral soils . Herein we find that integrating the dynamics of at a level of atmospheric methane abundance ([CH4]atm) of about HAMs and methanogens into a biogeochemistry model8–10 1.8 ppm (Fig. 1b)24. that includes permafrost soil organic carbon dynamics3 leads Quantification of the previously underestimated HAM-driven −1 to the upland methane sink doubling (~5.5 Tg CH4 yr ) north of methane sink is needed to improve our understanding of Arctic 50 °N in simulations from 2000–2016. The increase is equiva- methane budgets. -
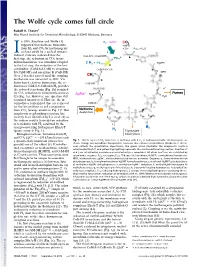
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Annotation Guidelines for Experimental Procedures
Annotation Guidelines for Experimental Procedures Developed By Mohammed Alliheedi Robert Mercer Version 1 April 14th, 2018 1- Introduction and background information What is rhetorical move? A rhetorical move can be defined as a text fragment that conveys a distinct communicative goal, in other words, a sentence that implies an author’s specific purpose to readers. What are the types of rhetorical moves? There are several types of rhetorical moves. However, we are interested in 4 rhetorical moves that are common in the method section of a scientific article that follows the Introduction Methods Results and Discussion (IMRaD) structure. 1- Description of a method: It is concerned with a sentence(s) that describes experimental events (e.g., “Beads with bound proteins were washed six times (for 10 min under rotation at 4°C) with pulldown buffer and proteins harvested in SDS-sample buffer, separated by SDS-PAGE, and analyzed by autoradiography.” (Ester & Uetz, 2008)). 2- Appeal to authority: It is concerned with a sentence(s) that discusses the use of standard methods, protocols, and procedures. There are two types of this move: - A reference to a well-established “standard” method (e.g., the use of a method like “PCR” or “electrophoresis”). - A reference to a method that was previously described in the literature (e.g., “Protein was determined using fluorescamine assay [41].” (Larsen, Frandesn and Treiman, 2001)). 3- Source of materials: It is concerned with a sentence(s) that lists the source of biological materials that are used in the experiment (e.g., “All microalgal strains used in this study are available at the Elizabeth Aidar Microalgae Culture Collection, Department of Marine Biology, Federal Fluminense University, Brazil.” (Larsen, Frandesn and Treiman, 2001)). -

Untargetted Metabolomic Exploration of the Mycobacterium Tuberculosis Stress Response to Cinnamon Essential Oil
biomolecules Article Untargetted Metabolomic Exploration of the Mycobacterium tuberculosis Stress Response to Cinnamon Essential Oil Elwira Sieniawska 1,* , Rafał Sawicki 2 , Joanna Golus 2 and Milen I. Georgiev 3,4 1 Chair and Department of Pharmacognosy, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland 2 Chair and Department of Biochemistry and Biotechnology, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland; [email protected] (R.S.); [email protected] (J.G.) 3 Group of Plant Cell Biotechnology and Metabolomics, The Stephan Angeloff Institute of Microbiology, Bulgarian Academy of Sciences, 139 Ruski Blvd., 4000 Plovdiv, Bulgaria; [email protected] 4 Center of Plant Systems Biology and Biotechnology, 4000 Plovdiv, Bulgaria * Correspondence: [email protected] Received: 6 January 2020; Accepted: 24 February 2020; Published: 26 February 2020 Abstract: The antimycobacterial activity of cinnamaldehyde has already been proven for laboratory strains and for clinical isolates. What is more, cinnamaldehyde was shown to threaten the mycobacterial plasma membrane integrity and to activate the stress response system. Following promising applications of metabolomics in drug discovery and development we aimed to explore the mycobacteria response to cinnamaldehyde within cinnamon essential oil treatment by untargeted liquid chromatography–mass spectrometry. The use of predictive metabolite pathway analysis and description of produced lipids enabled the evaluation of the stress symptoms shown by bacteria. This study suggests that bacteria exposed to cinnamaldehyde could reorganize their outer membrane as a physical barrier against stress factors. They probably lowered cell wall permeability and inner membrane fluidity, and possibly redirected carbon flow to store energy in triacylglycerols. Being a reactive compound, cinnamaldehyde may also contribute to disturbances in bacteria redox homeostasis and detoxification mechanisms. -

Towards Kinetic Modeling of Global Metabolic Networks with Incomplete Experimental Input on Kinetic Parameters
Towards Kinetic Modeling of Global Metabolic Networks with Incomplete Experimental Input on Kinetic Parameters Ping Ao 1,2 , Lik Wee Lee 1, Mary E. Lidstrom 3,4 , Lan Yin 5, and Xiaomei Zhu 6 1 Department of Mechanical Engineering , University of Washington , Seattle , WA 98195, USA 2 Department of Physics , University of Washington , Seattle , WA 98195, USA 3 Department of Microbiology , University of Washington , Seattle , WA 98195, USA 4 Department of Chemical Engineering , University of Washington , Seattle , WA 98195, USA 5 School of Physics , Peking University , 100871 Beijing , PR China 6 GeneMath , 5525 27 th Ave . N .E., Seattle , WA 98105, USA Abstract: This is the first report on a systematic method for constructing a large scale kinetic metabolic model and its initial application to the modeling of central metabolism of Methylobacterium extorquens AM1, a methylotrophic and environmental important bacterium. Its central metabolic network includes formaldehyde metabolism, serine cycle, citric acid cycle, pentose phosphate pathway, gluconeogensis, PHB synthesis and acetyl-CoA conversion pathway, respiration and energy metabolism. Through a systematic and consistent procedure of finding a set of parameters in the physiological range we overcome an outstanding difficulty in large scale kinetic modeling: the requirement for a massive number of enzymatic reaction parameters. We are able to construct the kinetic model based on general biological considerations and incomplete experimental kinetic parameters. Our method consists of -

The Ratio of Methanogens to Methanotrophs and Water-Level Dynamics Drive Methane Exchange Velocity in a Temperate Kettle-Hole Peat Bog
Biogeosciences Discuss., https://doi.org/10.5194/bg-2019-116 Manuscript under review for journal Biogeosciences Discussion started: 23 April 2019 c Author(s) 2019. CC BY 4.0 License. The ratio of methanogens to methanotrophs and water-level dynamics drive methane exchange velocity in a temperate kettle-hole peat bog Camilo Rey-Sanchez1,4, Gil Bohrer1, Julie Slater2, Yueh-Fen Li3, Roger Grau-Andrés2, Yushan Hao2, 5 Virginia I. Rich3, & G. Matt Davies2 1Department of Civil and Environmental Engineering and Geodetic Science, The Ohio State University, Columbus, Ohio, 43210, USA 2School of Environment and Natural Resources, The Ohio State University, Columbus, Ohio, 43210, USA 3Department of Microbiology, The Ohio State University, Columbus, Ohio, 43210, USA 10 4Current address, Department of Environmental Science, Management and Policy, University of California- Berkeley, California, 94720, USA Correspondence to: Camilo Rey-Sanchez ([email protected]) 15 Abstract. Peatlands are a large source of methane (CH4) to the atmosphere, yet the uncertainty around the estimates of CH4 flux from peatlands is large. To better understand the spatial heterogeneity in temperate peatland CH4 emissions and their response to physical and biological drivers, we studied CH4 dynamics throughout the growing seasons of 2017 and 2018 in Flatiron Lake Bog, a kettle-hole peat bog in Ohio. The site is composed of six different hydro-biological zones: an open water zone, four concentric vegetation zones surrounding the open water, and a restored zone connected to the main bog by a narrow 20 channel. At each of these locations, we monitored water level (WL), CH4 pore-water concentration at different peat depths, CH4 fluxes from the ground and from representative plant species using chambers, and microbial community composition with focus here on known methanogens and methanotrophs. -

Recycling of Vitamin B12 and NAD+ Within the Pdu Microcompartment of Salmonella Enterica Shouqiang Cheng Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2010 Recycling of vitamin B12 and NAD+ within the Pdu microcompartment of Salmonella enterica Shouqiang Cheng Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Biochemistry, Biophysics, and Structural Biology Commons Recommended Citation Cheng, Shouqiang, "Recycling of vitamin B12 and NAD+ within the Pdu microcompartment of Salmonella enterica" (2010). Graduate Theses and Dissertations. 11713. https://lib.dr.iastate.edu/etd/11713 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. + Recycling of vitamin B12 and NAD within the Pdu microcompartment of Salmonella enterica by Shouqiang Cheng A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Biochemistry Program of Study Committee: Thomas A. Bobik, Major Professor Alan DiSpirito Basil Nikolau Reuben Peters Gregory J. Phillips Iowa State University Ames, Iowa 2010 Copyright © Shouqiang Cheng, 2010. All rights reserved. ii Table of contents Abstract............................................................................................................................. -

Community and Proteomic Analysis of Anaerobic Consortia Converting Tetramethylammonium to Methane
Supporting Information for Manuscript Submitted to Archaea Community and Proteomic Analysis of Anaerobic Consortia Converting Tetramethylammonium to Methane Wei-Yu Chen1, Lucia Kraková2, Jer-Horng Wu1*, Domenico Pangallo2, Lenka Jeszeová2, Bing Liu3, Hidenari Yasui3 , 1Department of Environmental Engineering, National Cheng Kung University, No.1, University Road, East District, Tainan City 701, Taiwan (ROC) 2Institute of Molecular Biology, Slovak Academy of Sciences, Dubravska cesta 21, 84551 Bratislava, Slovakia 3Faculty of Environmental Engineering, The University of Kitakyushu, 1-1, Hibikino, Wakamatsu, Kitakyushu, Japan 808-0135 1 Table S1 Annotation of proteins related to the conversion of methylamines by the methanogens in the CMJP sample analyzed in this study. Relative Coverage Peptides Accession Organism gene Annotation abundance (%) (#) (%) A0A0E3SIM3 Methanosarcina barkeri 3 MSBR3_0618 Dimethylamine:corrinoid methyltransferase 17.13 21 1.39 A0A0E3Q5F5 Methanosarcina vacuolata Z-761 MSVAZ_1366 Methanol methyltransferase corrinoid protein 12.94 15 0.28 A0A0E3P0I6 Methanosarcina siciliae T4/M MSSIT_0113 Monomethylamine:corrinoid methyltransferase 23.58 20 0.48 A0A0E3WTL8 Methanosarcina lacustris Z-7289 MSLAZ_3204 Monomethylamine:corrinoid methyltransferase 15.94 9 0.24 A0A0E3KRC1 Methanosarcina thermophila CHTI-55 MSTHC_1357 Monomethylamine:corrinoid methyltransferase 18.12 10 0.22 A0A0E3WT78 Methanosarcina lacustris Z-7289 MSLAZ_2479 Dimethylamine methyltransferase corrinoid protein 25.00 11 0.13 A0A0E3Q3C8 Methanosarcina vacuolata -

(Antarctica) Glacial, Basal, and Accretion Ice
CHARACTERIZATION OF ORGANISMS IN VOSTOK (ANTARCTICA) GLACIAL, BASAL, AND ACCRETION ICE Colby J. Gura A Thesis Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE December 2019 Committee: Scott O. Rogers, Advisor Helen Michaels Paul Morris © 2019 Colby Gura All Rights Reserved iii ABSTRACT Scott O. Rogers, Advisor Chapter 1: Lake Vostok is named for the nearby Vostok Station located at 78°28’S, 106°48’E and at an elevation of 3,488 m. The lake is covered by a glacier that is approximately 4 km thick and comprised of 4 different types of ice: meteoric, basal, type 1 accretion ice, and type 2 accretion ice. Six samples were derived from the glacial, basal, and accretion ice of the 5G ice core (depths of 2,149 m; 3,501 m; 3,520 m; 3,540 m; 3,569 m; and 3,585 m) and prepared through several processes. The RNA and DNA were extracted from ultracentrifugally concentrated meltwater samples. From the extracted RNA, cDNA was synthesized so the samples could be further manipulated. Both the cDNA and the DNA were amplified through polymerase chain reaction. Ion Torrent primers were attached to the DNA and cDNA and then prepared to be sequenced. Following sequencing the sequences were analyzed using BLAST. Python and Biopython were then used to collect more data and organize the data for manual curation and analysis. Chapter 2: As a result of the glacier and its geographic location, Lake Vostok is an extreme and unique environment that is often compared to Jupiter’s ice-covered moon, Europa. -

Nitrification 31
NITROGEN IN SOILS/Nitrification 31 See also: Eutrophication; Greenhouse Gas Emis- Powlson DS (1993) Understanding the soil nitrogen cycle. sions; Isotopes in Soil and Plant Investigations; Soil Use and Management 9: 86–94. Nitrogen in Soils: Cycle; Nitrification; Plant Uptake; Powlson DS (1999) Fate of nitrogen from manufactured Symbiotic Fixation; Pollution: Groundwater fertilizers in agriculture. In: Wilson WS, Ball AS, and Hinton RH (eds) Managing Risks of Nitrates to Humans Further Reading and the Environment, pp. 42–57. Cambridge: Royal Society of Chemistry. Addiscott TM, Whitmore AP, and Powlson DS (1991) Powlson DS (1997) Integrating agricultural nutrient man- Farming, Fertilizers and the Nitrate Problem. Wallingford: agement with environmental objectives – current state CAB International. and future prospects. Proceedings No. 402. York: The Benjamin N (2000) Nitrates in the human diet – good or Fertiliser Society. bad? Annales de Zootechnologie 49: 207–216. Powlson DS, Hart PBS, Poulton PR, Johnston AE, and Catt JA et al. (1998) Strategies to decrease nitrate leaching Jenkinson DS (1986) Recovery of 15N-labelled fertilizer in the Brimstone Farm experiment, Oxfordshire, UK, applied in autumn to winter wheat at four sites in eastern 1988–1993: the effects of winter cover crops and England. Journal of Agricultural Science, Cambridge unfertilized grass leys. Plant and Soil 203: 57–69. 107: 611–620. Cheney K (1990) Effect of nitrogen fertilizer rate on soil Recous S, Fresnau C, Faurie G, and Mary B (1988) The fate nitrate nitrogen content after harvesting winter wheat. of labelled 15N urea and ammonium nitrate applied to a Journal of Agricultural Science, Cambridge 114: winter wheat crop.