Community and Proteomic Analysis of Anaerobic Consortia Converting Tetramethylammonium to Methane
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -
The Wolfe Cycle Comes Full Circle
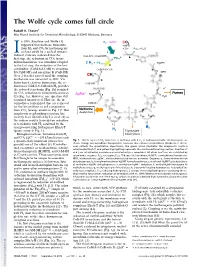
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Annotation Guidelines for Experimental Procedures
Annotation Guidelines for Experimental Procedures Developed By Mohammed Alliheedi Robert Mercer Version 1 April 14th, 2018 1- Introduction and background information What is rhetorical move? A rhetorical move can be defined as a text fragment that conveys a distinct communicative goal, in other words, a sentence that implies an author’s specific purpose to readers. What are the types of rhetorical moves? There are several types of rhetorical moves. However, we are interested in 4 rhetorical moves that are common in the method section of a scientific article that follows the Introduction Methods Results and Discussion (IMRaD) structure. 1- Description of a method: It is concerned with a sentence(s) that describes experimental events (e.g., “Beads with bound proteins were washed six times (for 10 min under rotation at 4°C) with pulldown buffer and proteins harvested in SDS-sample buffer, separated by SDS-PAGE, and analyzed by autoradiography.” (Ester & Uetz, 2008)). 2- Appeal to authority: It is concerned with a sentence(s) that discusses the use of standard methods, protocols, and procedures. There are two types of this move: - A reference to a well-established “standard” method (e.g., the use of a method like “PCR” or “electrophoresis”). - A reference to a method that was previously described in the literature (e.g., “Protein was determined using fluorescamine assay [41].” (Larsen, Frandesn and Treiman, 2001)). 3- Source of materials: It is concerned with a sentence(s) that lists the source of biological materials that are used in the experiment (e.g., “All microalgal strains used in this study are available at the Elizabeth Aidar Microalgae Culture Collection, Department of Marine Biology, Federal Fluminense University, Brazil.” (Larsen, Frandesn and Treiman, 2001)). -

Untargetted Metabolomic Exploration of the Mycobacterium Tuberculosis Stress Response to Cinnamon Essential Oil
biomolecules Article Untargetted Metabolomic Exploration of the Mycobacterium tuberculosis Stress Response to Cinnamon Essential Oil Elwira Sieniawska 1,* , Rafał Sawicki 2 , Joanna Golus 2 and Milen I. Georgiev 3,4 1 Chair and Department of Pharmacognosy, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland 2 Chair and Department of Biochemistry and Biotechnology, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland; [email protected] (R.S.); [email protected] (J.G.) 3 Group of Plant Cell Biotechnology and Metabolomics, The Stephan Angeloff Institute of Microbiology, Bulgarian Academy of Sciences, 139 Ruski Blvd., 4000 Plovdiv, Bulgaria; [email protected] 4 Center of Plant Systems Biology and Biotechnology, 4000 Plovdiv, Bulgaria * Correspondence: [email protected] Received: 6 January 2020; Accepted: 24 February 2020; Published: 26 February 2020 Abstract: The antimycobacterial activity of cinnamaldehyde has already been proven for laboratory strains and for clinical isolates. What is more, cinnamaldehyde was shown to threaten the mycobacterial plasma membrane integrity and to activate the stress response system. Following promising applications of metabolomics in drug discovery and development we aimed to explore the mycobacteria response to cinnamaldehyde within cinnamon essential oil treatment by untargeted liquid chromatography–mass spectrometry. The use of predictive metabolite pathway analysis and description of produced lipids enabled the evaluation of the stress symptoms shown by bacteria. This study suggests that bacteria exposed to cinnamaldehyde could reorganize their outer membrane as a physical barrier against stress factors. They probably lowered cell wall permeability and inner membrane fluidity, and possibly redirected carbon flow to store energy in triacylglycerols. Being a reactive compound, cinnamaldehyde may also contribute to disturbances in bacteria redox homeostasis and detoxification mechanisms. -

Towards Kinetic Modeling of Global Metabolic Networks with Incomplete Experimental Input on Kinetic Parameters
Towards Kinetic Modeling of Global Metabolic Networks with Incomplete Experimental Input on Kinetic Parameters Ping Ao 1,2 , Lik Wee Lee 1, Mary E. Lidstrom 3,4 , Lan Yin 5, and Xiaomei Zhu 6 1 Department of Mechanical Engineering , University of Washington , Seattle , WA 98195, USA 2 Department of Physics , University of Washington , Seattle , WA 98195, USA 3 Department of Microbiology , University of Washington , Seattle , WA 98195, USA 4 Department of Chemical Engineering , University of Washington , Seattle , WA 98195, USA 5 School of Physics , Peking University , 100871 Beijing , PR China 6 GeneMath , 5525 27 th Ave . N .E., Seattle , WA 98105, USA Abstract: This is the first report on a systematic method for constructing a large scale kinetic metabolic model and its initial application to the modeling of central metabolism of Methylobacterium extorquens AM1, a methylotrophic and environmental important bacterium. Its central metabolic network includes formaldehyde metabolism, serine cycle, citric acid cycle, pentose phosphate pathway, gluconeogensis, PHB synthesis and acetyl-CoA conversion pathway, respiration and energy metabolism. Through a systematic and consistent procedure of finding a set of parameters in the physiological range we overcome an outstanding difficulty in large scale kinetic modeling: the requirement for a massive number of enzymatic reaction parameters. We are able to construct the kinetic model based on general biological considerations and incomplete experimental kinetic parameters. Our method consists of -

Recycling of Vitamin B12 and NAD+ Within the Pdu Microcompartment of Salmonella Enterica Shouqiang Cheng Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2010 Recycling of vitamin B12 and NAD+ within the Pdu microcompartment of Salmonella enterica Shouqiang Cheng Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Biochemistry, Biophysics, and Structural Biology Commons Recommended Citation Cheng, Shouqiang, "Recycling of vitamin B12 and NAD+ within the Pdu microcompartment of Salmonella enterica" (2010). Graduate Theses and Dissertations. 11713. https://lib.dr.iastate.edu/etd/11713 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. + Recycling of vitamin B12 and NAD within the Pdu microcompartment of Salmonella enterica by Shouqiang Cheng A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Biochemistry Program of Study Committee: Thomas A. Bobik, Major Professor Alan DiSpirito Basil Nikolau Reuben Peters Gregory J. Phillips Iowa State University Ames, Iowa 2010 Copyright © Shouqiang Cheng, 2010. All rights reserved. ii Table of contents Abstract............................................................................................................................. -

Methane Utilization in Methylomicrobium Alcaliphilum 20ZR: a Systems Approach Received: 8 September 2017 Ilya R
www.nature.com/scientificreports OPEN Methane utilization in Methylomicrobium alcaliphilum 20ZR: a systems approach Received: 8 September 2017 Ilya R. Akberdin1,2, Merlin Thompson1, Richard Hamilton1, Nalini Desai3, Danny Alexander3, Accepted: 22 January 2018 Calvin A. Henard4, Michael T. Guarnieri4 & Marina G. Kalyuzhnaya1 Published: xx xx xxxx Biological methane utilization, one of the main sinks of the greenhouse gas in nature, represents an attractive platform for production of fuels and value-added chemicals. Despite the progress made in our understanding of the individual parts of methane utilization, our knowledge of how the whole-cell metabolic network is organized and coordinated is limited. Attractive growth and methane-conversion rates, a complete and expert-annotated genome sequence, as well as large enzymatic, 13C-labeling, and transcriptomic datasets make Methylomicrobium alcaliphilum 20ZR an exceptional model system for investigating methane utilization networks. Here we present a comprehensive metabolic framework of methane and methanol utilization in M. alcaliphilum 20ZR. A set of novel metabolic reactions governing carbon distribution across central pathways in methanotrophic bacteria was predicted by in-silico simulations and confrmed by global non-targeted metabolomics and enzymatic evidences. Our data highlight the importance of substitution of ATP-linked steps with PPi-dependent reactions and support the presence of a carbon shunt from acetyl-CoA to the pentose-phosphate pathway and highly branched TCA cycle. The diverged TCA reactions promote balance between anabolic reactions and redox demands. The computational framework of C1-metabolism in methanotrophic bacteria can represent an efcient tool for metabolic engineering or ecosystem modeling. Climate change concerns linked to increasing concentrations of anthropogenic methane have spiked inter- est in biological methane utilization as a novel platform for improving ecological and human well-being1–4. -

The Genome Sequence of Methanohalophilus Mahii SLPT
Hindawi Publishing Corporation Archaea Volume 2010, Article ID 690737, 16 pages doi:10.1155/2010/690737 Research Article TheGenomeSequenceofMethanohalophilus mahii SLPT Reveals Differences in the Energy Metabolism among Members of the Methanosarcinaceae Inhabiting Freshwater and Saline Environments Stefan Spring,1 Carmen Scheuner,1 Alla Lapidus,2 Susan Lucas,2 Tijana Glavina Del Rio,2 Hope Tice,2 Alex Copeland,2 Jan-Fang Cheng,2 Feng Chen,2 Matt Nolan,2 Elizabeth Saunders,2, 3 Sam Pitluck,2 Konstantinos Liolios,2 Natalia Ivanova,2 Konstantinos Mavromatis,2 Athanasios Lykidis,2 Amrita Pati,2 Amy Chen,4 Krishna Palaniappan,4 Miriam Land,2, 5 Loren Hauser,2, 5 Yun-Juan Chang,2, 5 Cynthia D. Jeffries,2, 5 Lynne Goodwin,2, 3 John C. Detter,3 Thomas Brettin,3 Manfred Rohde,6 Markus Goker,¨ 1 Tanja Woyke, 2 Jim Bristow,2 Jonathan A. Eisen,2, 7 Victor Markowitz,4 Philip Hugenholtz,2 Nikos C. Kyrpides,2 and Hans-Peter Klenk1 1 DSMZ—German Collection of Microorganisms and Cell Cultures GmbH, 38124 Braunschweig, Germany 2 DOE Joint Genome Institute, Walnut Creek, CA 94598-1632, USA 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, NM 87545-001, USA 4 Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA 5 Oak Ridge National Laboratory, Oak Ridge, TN 37830-8026, USA 6 HZI—Helmholtz Centre for Infection Research, 38124 Braunschweig, Germany 7 Davis Genome Center, University of California, Davis, CA 95817, USA Correspondence should be addressed to Stefan Spring, [email protected] and Hans-Peter Klenk, [email protected] Received 24 August 2010; Accepted 9 November 2010 Academic Editor: Valerie´ de Crecy-Lagard´ Copyright © 2010 Stefan Spring et al. -

View PDF Version
RSC Advances View Article Online REVIEW View Journal | View Issue Guidance for engineering of synthetic methylotrophy based on methanol metabolism in Cite this: RSC Adv.,2017,7,4083 methylotrophy Wenming Zhang, Ting Zhang, Sihua Wu, Mingke Wu, Fengxue Xin, Weiliang Dong, Jiangfeng Ma, Min Zhang and Min Jiang* Methanol is increasingly becoming an attractive substrate for production of different metabolites, such as commodity chemicals, and biofuels via biological conversion, due to the increment of annual production capacity and decrement of prices. In recent years, genetic engineering towards native menthol utilizing organisms – methylotrophy has developed rapidly and attracted widespread attention. Therefore, it is vital to elucidate the distinct pathways that involve methanol oxidation, formaldehyde assimilation and disassimilation in the different methylotrophies for future synthetic work. In addition, this will also help to genetically construct some new and non-native methylotrophies. This review summarizes the Received 19th November 2016 Creative Commons Attribution-NonCommercial 3.0 Unported Licence. current knowledge about the methanol metabolism pathways in methylotrophy, discusses and Accepted 26th December 2016 compares different pathways on methanol utilization, and finally presents the strategies to integrate DOI: 10.1039/c6ra27038g the methanol metabolism with other chemicals, biofuels or other high value-added product formation www.rsc.org/advances pathways. 1. Introduction million tons per year and an expected annual growth rate in the range of 10–20%, a methanol-based bioeconomy has been With the rapid growth of the world population and develop- proposed.3 Especially recently, with the rise of the methanol This article is licensed under a ment of industry and society, energy demand is dramatically production process, the price of methanol steadily declined. -

Four Families of Folate-Independent Methionine Synthases Morgan N
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.21.054031; this version posted November 9, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Four Families of Folate-independent Methionine Synthases Morgan N. Price, Adam M. Deutschbauer, and Adam P. Arkin Lawrence Berkeley National Lab Abstract Although most organisms synthesize methionine from homocysteine and methyl folates, some have “core” methionine synthases that lack folate-binding domains and use other methyl donors. In vitro, the characterized core synthases use methylcobalamin as a methyl donor, but in vivo, they probably rely on corrinoid (vitamin B12-binding) proteins. We identified four families of core methionine synthases that are distantly related to each other (under 30% pairwise amino acid identity). From the characterized enzymes, we identified the families MesA, which is found in methanogens, and MesB, which is found in anaerobic bacteria and archaea with the Wood-Ljungdahl pathway. A third uncharacterized family, MesC, is found in anaerobic archaea that have the Wood-Ljungdahl pathway and lack known forms of methionine synthase. We predict that most members of the MesB and MesC families accept methyl groups from the iron-sulfur corrinoid protein of that pathway. The fourth family, MesD, is found only in aerobic bacteria. Using transposon mutants and complementation, we show that MesD does not require 5-methyltetrahydrofolate or cobalamin. Instead, MesD requires an uncharacterized protein family (DUF1852) and oxygen for activity. -

Lanthanide-Dependent Methanol and Formaldehyde Oxidation in Methylobacterium Aquaticum Strain 22A
microorganisms Article Lanthanide-Dependent Methanol and Formaldehyde Oxidation in Methylobacterium aquaticum Strain 22A Patcha Yanpirat 1, Yukari Nakatsuji 1, Shota Hiraga 1, Yoshiko Fujitani 1, Terumi Izumi 1, Sachiko Masuda 1,2,3, Ryoji Mitsui 4, Tomoyuki Nakagawa 5,6 and Akio Tani 1,* 1 Institute of Plant Science and Resources, Okayama University, Okayama 710-0046, Japan; [email protected] (P.Y.); [email protected] (Y.N.); [email protected] (S.H.); [email protected] (Y.F.); [email protected] (T.I.); [email protected] (S.M.) 2 Advanced Low Carbon Technology Research and Development Program, Japan Science and Technology Agency, Tokyo 102-0076, Japan 3 RIKEN Center for Sustainable Resource Science, Kanagawa 230-0045, Japan 4 Department of Biochemistry, Faculty of Science, Okayama University of Science, Okayama 700-8530, Japan; [email protected] 5 The United Graduate School of Agricultural Science, Gifu University, Gifu 501-1193, Japan; [email protected] 6 The Graduate School of Natural Sciences and Technologies, Gifu University, Gifu 501-1193, Japan * Correspondence: [email protected] Received: 5 May 2020; Accepted: 28 May 2020; Published: 30 May 2020 Abstract: Lanthanides (Ln) are an essential cofactor for XoxF-type methanol dehydrogenases (MDHs) in Gram-negative methylotrophs. The Ln3+ dependency of XoxF has expanded knowledge and raised new questions in methylotrophy, including the differences in characteristics of XoxF-type MDHs, their regulation, and the methylotrophic metabolism including formaldehyde oxidation. In this study, we genetically identified one set of Ln3+- and Ca2+-dependent MDHs (XoxF1 and MxaFI), that are involved in methylotrophy, and an ExaF-type Ln3+-dependent ethanol dehydrogenase, among six MDH-like genes in Methylobacterium aquaticum strain 22A. -

Characterization of Natural Product Biological Imprints for Computer-Aided Drug Design Applications Noe Sturm
Characterization of natural product biological imprints for computer-aided drug design applications Noe Sturm To cite this version: Noe Sturm. Characterization of natural product biological imprints for computer-aided drug design applications. Cheminformatics. Université de Strasbourg, 2015. English. NNT : 2015STRAF059. tel-01300872 HAL Id: tel-01300872 https://tel.archives-ouvertes.fr/tel-01300872 Submitted on 11 Apr 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITÉ DE STRASBOURG ÉCOLE DOCTORALE DES SCIENCES CHIMIQUES Laboratoire d’Innovation Thérapeutique, UMR 7200 en cotutelle avec Eskitis Institute for Drug Discovery, Griffith University THÈSE présentée par Noé STURM soutenue le : 8 Décembre 2015 pour obtenir le grade de : Docteur de l’université de Strasbourg Discipline/Spécialité : Chimie/Chémoinformatique Caractérisation de l’empreinte biologique des produits naturels pour des applications de conception rationnelle de médicament assistée par ordinateur THÈSE dirigée par : KELLENBERGER Esther Professeur, Université de Strasbourg QUINN Ronald Professeur, Université de Griffith, Brisbane, Australie RAPPORTEURS : IORGA Bogdan Chargé de recherche HDR, Institut de Chimie des Substances Naturelles, Gif-sur-Yvette GÜNTHER Stefan Professeur, Université Albert Ludwigs, Fribourg, Allemagne Acknowledgments First of all, I would like to thank my two supervisors Professor Kellenberger Esther and Professor Quinn Ronald for their excellent assistance, both academic and personal in nature.