Methylotrophic Methanogenesis in Hydraulically Fractured Shales
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Oxidative Pentose Phosphate Pathway Enzyme 6-Phosphogluconate Dehydrogenase Plays a Key Role in Breast Cancer Metabolism
biology Article Oxidative Pentose Phosphate Pathway Enzyme 6-Phosphogluconate Dehydrogenase Plays a Key Role in Breast Cancer Metabolism Ibrahim H. Polat 1,2,3 ,Míriam Tarrado-Castellarnau 1,4 , Rohit Bharat 1, Jordi Perarnau 1 , Adrian Benito 1,5 , Roldán Cortés 1, Philippe Sabatier 2 and Marta Cascante 1,4,* 1 Department of Biochemistry and Molecular Biomedicine and Institute of Biomedicine (IBUB), Faculty of Biology, Universitat de Barcelona, Av Diagonal 643, 08028 Barcelona, Spain; [email protected] (I.H.P.); [email protected] (M.T.-C.); [email protected] (R.B.); [email protected] (J.P.); [email protected] (A.B.); [email protected] (R.C.) 2 Equipe Environnement et Prédiction de la Santé des Populations, Laboratoire TIMC (UMR 5525), CHU de Grenoble, Université Grenoble Alpes, 38700 CEDEX La Tronche, France; [email protected] 3 Department of Medicine, Hematology/Oncology, Goethe-University Frankfurt, 60590 Frankfurt, Germany 4 Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBEREHD), Instituto de Salud Carlos III (ISCIII), 28001 Madrid, Spain 5 Division of Cancer, Department of Surgery and Cancer, Faculty of Medicine, Imperial College London, London W12 0NN, UK * Correspondence: [email protected] Simple Summary: Cancer cells alter their metabolism to maintain their high need for energy, produce Citation: Polat, I.H.; enough macromolecules for biosynthesis, and preserve their redox status. The investigation of Tarrado-Castellarnau, M.; Bharat, R.; cancer cell-specific metabolic alterations has vital importance to identify targets to be exploited for Perarnau, J.; Benito, A.; Cortés, R.; therapeutic development. The pentose phosphate pathway (PPP) is often highly activated in tumor Sabatier, P.; Cascante, M. -

Regeneration of Unconventional Natural Gas by Methanogens Co
www.nature.com/scientificreports OPEN Regeneration of unconventional natural gas by methanogens co‑existing with sulfate‑reducing prokaryotes in deep shale wells in China Yimeng Zhang1,2,3, Zhisheng Yu1*, Yiming Zhang4 & Hongxun Zhang1 Biogenic methane in shallow shale reservoirs has been proven to contribute to economic recovery of unconventional natural gas. However, whether the microbes inhabiting the deeper shale reservoirs at an average depth of 4.1 km and even co-occurring with sulfate-reducing prokaryote (SRP) have the potential to produce biomethane is still unclear. Stable isotopic technique with culture‑dependent and independent approaches were employed to investigate the microbial and functional diversity related to methanogenic pathways and explore the relationship between SRP and methanogens in the shales in the Sichuan Basin, China. Although stable isotopic ratios of the gas implied a thermogenic origin for methane, the decreased trend of stable carbon and hydrogen isotope value provided clues for increasing microbial activities along with sustained gas production in these wells. These deep shale-gas wells harbored high abundance of methanogens (17.2%) with ability of utilizing various substrates for methanogenesis, which co-existed with SRP (6.7%). All genes required for performing methylotrophic, hydrogenotrophic and acetoclastic methanogenesis were present. Methane production experiments of produced water, with and without additional available substrates for methanogens, further confrmed biomethane production via all three methanogenic pathways. Statistical analysis and incubation tests revealed the partnership between SRP and methanogens under in situ sulfate concentration (~ 9 mg/L). These results suggest that biomethane could be produced with more fexible stimulation strategies for unconventional natural gas recovery even at the higher depths and at the presence of SRP. -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

The Vertical Distribution of Sediment Archaeal Community in the (Black Bloom) Disturbing Zhushan Bay of Lake Taihu
Hindawi Publishing Corporation Archaea Volume 2016, Article ID 8232135, 8 pages http://dx.doi.org/10.1155/2016/8232135 Research Article The Vertical Distribution of Sediment Archaeal Community in the (Black Bloom) Disturbing Zhushan Bay of Lake Taihu Xianfang Fan1,2 and Peng Xing1 1 State Key Laboratory of Lake Science and Environment, Nanjing Institute of Geography & Limnology, Chinese Academy of Sciences, Nanjing 210008, China 2State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing 210008, China Correspondence should be addressed to Peng Xing; [email protected] Received 20 August 2015; Revised 27 November 2015; Accepted 20 December 2015 Academic Editor: William B. Whitman Copyright © 2016 X. Fan and P. Xing. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Using the Illumina sequencing technology, we investigated the vertical distribution of archaeal community in the sediment of Zhushan Bay of Lake Taihu, where the black bloom frequently occurred in summer. Overall, the Miscellaneous Crenarchaeotal Group (MCG), Deep Sea Hydrothermal Vent Group 6 (DHVEG-6), and Methanobacterium dominated the archaeal community. However, we observed significant difference in composition of archaeal community among different depths of the sediment. DHVEG-6 dominated in the surface layer (0–3 cm) sediment. Methanobacterium was the dominating archaeal taxa in the L2 (3– 6 cm) and L3 (6–10) sediment. MCG was most abundant in the L4 (10–15 cm) and L5 (15–20 cm) sediment. Besides, DHVEG-6 was significantly affected by the concentration of total phosphorus ).(TP And loss on ignition (LOI) was an important environmental factor for Methanobacterium. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Enzyme Characterisation and Kinetic Modelling of the Pentose Phosphate
1 Enzyme characterisation and kinetic modelling of the pentose 2 phosphate pathway in yeast 1;2 3 Hanan L. Messiha Edward Kent 1;3;4 Naglis Malys 1;2;6 Kathleen M. Carroll 1;5 Neil Swainston 1;4 s 1;4;7;∗ t Pedro Mendes 1;4 n Kieran Smallbone i r P 1Manchester Centre for Integrative Systems Biology e r 2Faculty of Life Sciences 3 P Doctoral Training Centre in Integrative Systems Biology 4School of Computer Science 5School of Chemistry University of Manchester, M13 9PL, UK. 6School of Life Sciences, Gibbet Hill Campus, University of Warwick, Coventry, UK. 7Center for Quantitative Medicine and Department of Cell Biology, University of Connecticut Health Center, 263 Farmington Avenue, Farmington, CT 06030, USA. 4 Abstract 5 We present the quantification and kinetic characterisation of the enzymes of the pentose 6 phosphate pathway in Saccharomyces cerevisiae. The data are combined into a mathematical 7 model that describes the dynamics of this system and allows us to predict changes in metabo- 8 lite concentrations and fluxes in response to perturbations. We use the model to study the 9 response of yeast to a glucose pulse. We then combine the model with an existing glycolysis 10 model to study the effect of oxidative stress on carbohydrate metabolism. The combina- 11 tion of these two models was made possible by the standardised enzyme kinetic experiments 12 carried out in both studies. This work demonstrates the feasibility of constructing larger 13 network-scale models by merging smaller pathway-scale models. ∗To whom correspondence should be addressed at [email protected] PeerJ PrePrints | http://dx.doi.org/10.7287/peerj.preprints.146v4 | CC-BY 3.0 Open Access | received: 10 Apr 2014, published: 10 Apr 2014 1 14 Introduction 15 The pentose phosphate pathway (PPP) is a central and widely conserved metabolic pathway of car- 16 bohydrate metabolism which, in eukaryotic cells, is located in the cytoplasm (see Figure 1). -

Cryptic Inoviruses Revealed As Pervasive in Bacteria and Archaea Across Earth’S Biomes
ARTICLES https://doi.org/10.1038/s41564-019-0510-x Corrected: Author Correction Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes Simon Roux 1*, Mart Krupovic 2, Rebecca A. Daly3, Adair L. Borges4, Stephen Nayfach1, Frederik Schulz 1, Allison Sharrar5, Paula B. Matheus Carnevali 5, Jan-Fang Cheng1, Natalia N. Ivanova 1, Joseph Bondy-Denomy4,6, Kelly C. Wrighton3, Tanja Woyke 1, Axel Visel 1, Nikos C. Kyrpides1 and Emiley A. Eloe-Fadrosh 1* Bacteriophages from the Inoviridae family (inoviruses) are characterized by their unique morphology, genome content and infection cycle. One of the most striking features of inoviruses is their ability to establish a chronic infection whereby the viral genome resides within the cell in either an exclusively episomal state or integrated into the host chromosome and virions are continuously released without killing the host. To date, a relatively small number of inovirus isolates have been extensively studied, either for biotechnological applications, such as phage display, or because of their effect on the toxicity of known bacterial pathogens including Vibrio cholerae and Neisseria meningitidis. Here, we show that the current 56 members of the Inoviridae family represent a minute fraction of a highly diverse group of inoviruses. Using a machine learning approach lever- aging a combination of marker gene and genome features, we identified 10,295 inovirus-like sequences from microbial genomes and metagenomes. Collectively, our results call for reclassification of the current Inoviridae family into a viral order including six distinct proposed families associated with nearly all bacterial phyla across virtually every ecosystem. -
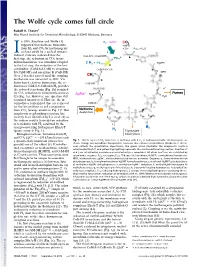
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

THE MASS of L-PYRROLYSINE in METHYLAMINE METHYLTRANSFERASES and the ROLE of ITS IMINE BOND in CATALYSIS DISSERTATION Presented I
THE MASS OF L-PYRROLYSINE IN METHYLAMINE METHYLTRANSFERASES AND THE ROLE OF ITS IMINE BOND IN CATALYSIS DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Graduate School of The Ohio State University by Jitesh Anthony Aloysius Soares, M.S. The Ohio State University 2008 Dissertation Committee: Dr. Joseph A. Krzycki, Advisor Approved by Dr. Charles J. Daniels Dr. Mark Morrison ______________________ Dr. F. Robert Tabita Advisor Graduate Program in Microbiology ABSTRACT Methanosarcina barkeri is an archaeon capable of producing methane from methylamines. Methylamine methyltransferases initiate methanogenesis from methylamines by transferring methyl groups to a cognate corrinoid protein. Each gene encoding a methylamine methyltransferase has been shown to contain a single in-frame amber codon. Further studies have shown that in the monomethylamine methyltransferase, mtmB , the amber codon encodes a novel amino acid, L-pyrrolysine. X-ray crystal structures of MtmB have shown that the structure of this amino acid is a lysine residue with the epsilon-nitrogen in amide linkage to a (4R, 5R)-4-substituted pyrrolyine-5-carboxylate ring. However, these structures did not allow an assignment of the pyrroline ring C4 substituent as a methyl or amine group. In this thesis (Chapter 2) mass spectrometry of chymotryptic digests of methylamine methyltransferases is employed to show that pyrrolysine in present in all three types of methylamine methyltransferase at the position corresponding to the amber codon in their respective genes. The mass of this amber-encoded residue was observed to coincide with the predicted mass of pyrrolysine with a methyl- group at the C4 position. -

Isolation and Characterization of Methanohalophilus Portucalensis Sp
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, July 1993, p. 430-437 Vol. 43, No. 3 0020-7713/93/030430-08$02.00/0 Copyright 0 1993, International Union of Microbiological Societies Isolation and Characterization of Methanohalophilus portucalensis sp. nov. and DNA Reassociation Study of the Genus Methanohalophilus DAVID R. BOONE,ly2* INDRA M. MATHRLWI,~?YITAT LIU,l JOSE A. G. F. MENAIA,1-4 ROBERT A. AND JANE E. BOONE' Departments of Environmental Science and Engineering' and Chemical and Biological Sciences, Oregon Graduate Institute of Science & Technology, 19600 N. W. von Neumann Drive, Beaverton, Oregon 97006- 1999; School of Public Health, University of California, Los Angeles, California 900243; and Laboratbrio Nacional de Engenharia e Tecnologia Industrial, Estrada das Palmeiras, 2745 Queluz, Portugal4 Six strains of coccoid, halophilic methanogens were isolated from various salinaria and natural hypersaline environments. These isolates (strains FDF-lT [T = type strain], FDF-2, SF-2, Ret-1, SD-1, and Cas-1) grew on media containing methanol and mono-, di-, and trimethylamines as catabolic substrates, but not on media containing dimethyl sulfide, methane thiol, H,, formate, or acetate; when cells were provided with H, in addition to methanol or trimethylamine, they grew on the medium containing a methyl substrate but did not catabolize H,. All of the strains were capable of growth in mineral medium to which trimethylamine was added as a catabolic substrate, although some strains were greatly stimulated by biotin or p-aminobenzoate. DNA reassociation and denaturing electrophoresis of whole-cell proteins indicated that strains FDF-lT, FDF-2, SF-2, and Ret-1, together with previously described strains SF-1, 2-7302, 2-7401, 2-7404, and 2-7405, belong to a new taxon named Methunohalophilzuportucalensis sp. -

Archaeology of Eukaryotic DNA Replication
Downloaded from http://cshperspectives.cshlp.org/ on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Archaeology of Eukaryotic DNA Replication Kira S. Makarova and Eugene V. Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894 Correspondence: [email protected] Recent advances in the characterization of the archaeal DNA replication system together with comparative genomic analysis have led to the identification of several previously un- characterized archaeal proteins involved in replication and currently reveal a nearly com- plete correspondence between the components of the archaeal and eukaryotic replication machineries. It can be inferred that the archaeal ancestor of eukaryotes and even the last common ancestor of all extant archaea possessed replication machineries that were compa- rable in complexity to the eukaryotic replication system. The eukaryotic replication system encompasses multiple paralogs of ancestral components such that heteromeric complexes in eukaryotes replace archaeal homomeric complexes, apparently along with subfunctionali- zation of the eukaryotic complex subunits. In the archaea, parallel, lineage-specific dupli- cations of many genes encoding replication machinery components are detectable as well; most of these archaeal paralogs remain to be functionally characterized. The archaeal rep- lication system shows remarkable plasticity whereby even some essential components such as DNA polymerase and single-stranded DNA-binding protein are displaced by unrelated proteins with analogous activities in some lineages. ouble-stranded DNA is the molecule that Okazaki fragments (Kornberg and Baker 2005; Dcarries genetic information in all cellular Barry and Bell 2006; Hamdan and Richardson life-forms; thus, replication of this genetic ma- 2009; Hamdan and van Oijen 2010). -

Differences in Lateral Gene Transfer in Hypersaline Versus Thermal Environments Matthew E Rhodes1*, John R Spear2, Aharon Oren3 and Christopher H House1
Rhodes et al. BMC Evolutionary Biology 2011, 11:199 http://www.biomedcentral.com/1471-2148/11/199 RESEARCH ARTICLE Open Access Differences in lateral gene transfer in hypersaline versus thermal environments Matthew E Rhodes1*, John R Spear2, Aharon Oren3 and Christopher H House1 Abstract Background: The role of lateral gene transfer (LGT) in the evolution of microorganisms is only beginning to be understood. While most LGT events occur between closely related individuals, inter-phylum and inter-domain LGT events are not uncommon. These distant transfer events offer potentially greater fitness advantages and it is for this reason that these “long distance” LGT events may have significantly impacted the evolution of microbes. One mechanism driving distant LGT events is microbial transformation. Theoretically, transformative events can occur between any two species provided that the DNA of one enters the habitat of the other. Two categories of microorganisms that are well-known for LGT are the thermophiles and halophiles. Results: We identified potential inter-class LGT events into both a thermophilic class of Archaea (Thermoprotei) and a halophilic class of Archaea (Halobacteria). We then categorized these LGT genes as originating in thermophiles and halophiles respectively. While more than 68% of transfer events into Thermoprotei taxa originated in other thermophiles, less than 11% of transfer events into Halobacteria taxa originated in other halophiles. Conclusions: Our results suggest that there is a fundamental difference between LGT in thermophiles and halophiles. We theorize that the difference lies in the different natures of the environments. While DNA degrades rapidly in thermal environments due to temperature-driven denaturization, hypersaline environments are adept at preserving DNA.