Archaeology of Eukaryotic DNA Replication
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Complete Genome Sequence of the Antarctic Halorubrum Lacusprofundi Type Strain ACAM 34 Iain J
Anderson et al. Standards in Genomic Sciences (2016) 11:70 DOI 10.1186/s40793-016-0194-2 SHORT GENOME REPORT Open Access Complete genome sequence of the Antarctic Halorubrum lacusprofundi type strain ACAM 34 Iain J. Anderson1, Priya DasSarma2*, Susan Lucas1, Alex Copeland1, Alla Lapidus1, Tijana Glavina Del Rio1, Hope Tice1, Eileen Dalin1, David C. Bruce3, Lynne Goodwin3, Sam Pitluck1, David Sims3, Thomas S. Brettin3, John C. Detter3, Cliff S. Han3, Frank Larimer1,4, Loren Hauser1,4, Miriam Land1,4, Natalia Ivanova1, Paul Richardson1, Ricardo Cavicchioli5, Shiladitya DasSarma2, Carl R. Woese6 and Nikos C. Kyrpides1 Abstract Halorubrum lacusprofundi is an extreme halophile within the archaeal phylum Euryarchaeota. The type strain ACAM 34 was isolated from Deep Lake, Antarctica. H. lacusprofundi is of phylogenetic interest because it is distantly related to the haloarchaea that have previously been sequenced. It is also of interest because of its psychrotolerance. WereportherethecompletegenomesequenceofH. lacusprofundi type strain ACAM 34 and its annotation. This genome is part of a 2006 Joint Genome Institute Community Sequencing Program project to sequence genomes of diverse Archaea. Keywords: Archaea, Halophile, Halorubrum, Extremophile, Cold adaptation, Tree of life Abbreviations: TE, Tris-EDTA buffer; CRITICA, Coding region identification tool invoking comparative analysis; PRIAM, PRofils pour l’Identification Automatique du Métabolisme; KEGG, Kyoto Encyclopedia of Genes and Genomes; COG, Clusters of Orthologous Groups; TMHMM, Transmembrane hidden Markov model; CRISPR, Clustered regularly interspaced short palindromic repeats Introduction 2006 Joint Genome Institute Community Sequencing Halorubrum lacusprofundi is an extremely halophilic Program project because of its ability to grow at low archaeon belonging to the class Halobacteria within the temperature and its phylogenetic distance from other phylum Euryarchaeota. -

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide
microorganisms Article Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide Kelly J. Hidalgo 1,2,* , Isabel N. Sierra-Garcia 3 , German Zafra 4 and Valéria M. de Oliveira 1 1 Microbial Resources Division, Research Center for Chemistry, Biology and Agriculture (CPQBA), University of Campinas–UNICAMP, Av. Alexandre Cazellato 999, 13148-218 Paulínia, Brazil; [email protected] 2 Graduate Program in Genetics and Molecular Biology, Institute of Biology, University of Campinas (UNICAMP), Rua Monteiro Lobato 255, Cidade Universitária, 13083-862 Campinas, Brazil 3 Biology Department & CESAM, University of Aveiro, Aveiro, Portugal, Campus de Santiago, Avenida João Jacinto de Magalhães, 3810-193 Aveiro, Portugal; [email protected] 4 Grupo de Investigación en Bioquímica y Microbiología (GIBIM), Escuela de Microbiología, Universidad Industrial de Santander, Cra 27 calle 9, 680002 Bucaramanga, Colombia; [email protected] * Correspondence: [email protected]; Tel.: +55-19981721510 Abstract: Microorganisms inhabiting subsurface petroleum reservoirs are key players in biochemical transformations. The interactions of microbial communities in these environments are highly complex and still poorly understood. This work aimed to assess publicly available metagenomes from oil reservoirs and implement a robust pipeline of genome-resolved metagenomics to decipher metabolic and taxonomic profiles of petroleum reservoirs worldwide. Analysis of 301.2 Gb of metagenomic information derived from heavily flooded petroleum reservoirs in China and Alaska to non-flooded petroleum reservoirs in Brazil enabled us to reconstruct 148 metagenome-assembled genomes (MAGs) of high and medium quality. At the phylum level, 74% of MAGs belonged to bacteria and 26% to archaea. The profiles of these MAGs were related to the physicochemical parameters and recovery management applied. -

Marsarchaeota Are an Aerobic Archaeal Lineage Abundant in Geothermal Iron Oxide Microbial Mats
Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats Authors: Zackary J. Jay, Jacob P. Beam, Mansur Dlakic, Douglas B. Rusch, Mark A. Kozubal, and William P. Inskeep This is a postprint of an article that originally appeared in Nature Microbiology on May 14, 2018. The final version can be found at https://dx.doi.org/10.1038/s41564-018-0163-1. Jay, Zackary J. , Jacob P. Beam, Mensur Dlakic, Douglas B. Rusch, Mark A. Kozubal, and William P. Inskeep. "Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats." Nature Microbiology 3, no. 6 (May 2018): 732-740. DOI: 10.1038/ s41564-018-0163-1. Made available through Montana State University’s ScholarWorks scholarworks.montana.edu Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats Zackary J. Jay1,4,7, Jacob P. Beam1,5,7, Mensur Dlakić2, Douglas B. Rusch3, Mark A. Kozubal1,6 and William P. Inskeep 1* The discovery of archaeal lineages is critical to our understanding of the universal tree of life and evolutionary history of the Earth. Geochemically diverse thermal environments in Yellowstone National Park provide unprecedented opportunities for studying archaea in habitats that may represent analogues of early Earth. Here, we report the discovery and character- ization of a phylum-level archaeal lineage proposed and herein referred to as the ‘Marsarchaeota’, after the red planet. The Marsarchaeota contains at least two major subgroups prevalent in acidic, microaerobic geothermal Fe(III) oxide microbial mats across a temperature range from ~50–80 °C. Metagenomics, single-cell sequencing, enrichment culturing and in situ transcrip- tional analyses reveal their biogeochemical role as facultative aerobic chemoorganotrophs that may also mediate the reduction of Fe(III). -

Large-Scale Trna Intron Transposition in The
Large-Scale tRNA Intron Transposition in the Archaeal Order Thermoproteales Represents a Novel Mechanism of Intron Gain Kosuke Fujishima,1,2 Junichi Sugahara,1,3 Masaru Tomita,1,2,3 and Akio Kanai*,1,2,3 1Institute for Advanced Biosciences, Keio University, Tsuruoka, Japan 2Department of Environmental Information, Keio University, Fujisawa, Japan 3Systems Biology Program, Graduate School of Media and Governance, Keio University, Fujisawa, Japan *Corresponding author: E-mail: [email protected]. Associate editor: Martin Embley Abstract Research article Recently, diverse arrangements of transfer RNA (tRNA) genes have been found in the domain Archaea, in which the tRNA is interrupted by a maximum of three introns or is even fragmented into two or three genes. Whereas most of the eukaryotic tRNA introns are inserted strictly at the canonical nucleotide position (37/38), archaeal intron-containing tRNAs have a wide diversity of small tRNA introns, which differ in their numbers and locations. This feature is especially pronounced in the archaeal order Thermoproteales. In this study, we performed a comprehensive sequence comparison of 286 tRNA introns and their genes in seven Thermoproteales species to clarify how these introns have emerged and diversified during tRNA gene evolution. We identified 46 intron groups containing sets of highly similar sequences (.70%) and showed that 16 of them contain sequences from evolutionarily distinct tRNA genes. The phylogeny of these 16 intron groups indicates that transposition events have occurred at least seven times throughout the evolution of Thermoproteales. These findings suggest that frequent intron transposition occurs among the tRNA genes of Thermoproteales. Further computational analysis revealed limited insertion positions and corresponding amino acid types of tRNA genes. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

The Role of Polyphosphate in Motility, Adhesion, and Biofilm Formation in Sulfolobales. Microorganisms 2021, 9
microorganisms Article The Role of Polyphosphate in Motility, Adhesion, and Biofilm Formation in Sulfolobales Alejandra Recalde 1,2 , Marleen van Wolferen 2 , Shamphavi Sivabalasarma 2 , Sonja-Verena Albers 2, Claudio A. Navarro 1 and Carlos A. Jerez 1,* 1 Laboratory of Molecular Microbiology and Biotechnology, Department of Biology, Faculty of Sciences, University of Chile, Santiago 8320000, Chile; [email protected] (A.R.); [email protected] (C.A.N.) 2 Laboratory of Molecular Biology of Archaea, Institute of Biology II-Microbiology, University of Freiburg, 79085 Freiburg, Germany; [email protected] (M.v.W.); [email protected] (S.S.); [email protected] (S.-V.A.) * Correspondence: [email protected] Abstract: Polyphosphates (polyP) are polymers of orthophosphate residues linked by high-energy phosphoanhydride bonds that are important in all domains of life and function in many different processes, including biofilm development. To study the effect of polyP in archaeal biofilm formation, our previously described Sa. solfataricus polyP (−) strain and a new polyP (−) S. acidocaldarius strain generated in this report were used. These two strains lack the polymer due to the overexpression of their respective exopolyphosphatase gene (ppx). Both strains showed a reduction in biofilm formation, decreased motility on semi-solid plates and a diminished adherence to glass surfaces as seen by DAPI (40,6-diamidino-2-phenylindole) staining using fluorescence microscopy. Even though arlB (encoding the archaellum subunit) was highly upregulated in S. acidocardarius polyP (−), no archaellated cells were observed. These results suggest that polyP might be involved in the regulation of the expression of archaellum components and their assembly, possibly by affecting energy availability, phosphorylation or other phenomena. -

Yu-Chen Ling and John W. Moreau
Microbial Distribution and Activity in a Coastal Acid Sulfate Soil System Introduction: Bioremediation in Yu-Chen Ling and John W. Moreau coastal acid sulfate soil systems Method A Coastal acid sulfate soil (CASS) systems were School of Earth Sciences, University of Melbourne, Melbourne, VIC 3010, Australia formed when people drained the coastal area Microbial distribution controlled by environmental parameters Microbial activity showed two patterns exposing the soil to the air. Drainage makes iron Microbial structures can be grouped into three zones based on the highest similarity between samples (Fig. 4). Abundant populations, such as Deltaproteobacteria, kept constant activity across tidal cycling, whereas rare sulfides oxidize and release acidity to the These three zones were consistent with their geological background (Fig. 5). Zone 1: Organic horizon, had the populations changed activity response to environmental variations. Activity = cDNA/DNA environment, low pH pore water further dissolved lowest pH value. Zone 2: surface tidal zone, was influenced the most by tidal activity. Zone 3: Sulfuric zone, Abundant populations: the heavy metals. The acidity and toxic metals then Method A Deltaproteobacteria Deltaproteobacteria this area got neutralized the most. contaminate coastal and nearby ecosystems and Method B 1.5 cause environmental problems, such as fish kills, 1.5 decreased rice yields, release of greenhouse gases, Chloroflexi and construction damage. In Australia, there is Gammaproteobacteria Gammaproteobacteria about a $10 billion “legacy” from acid sulfate soils, Chloroflexi even though Australia is only occupied by around 1.0 1.0 Cyanobacteria,@ Acidobacteria Acidobacteria Alphaproteobacteria 18% of the global acid sulfate soils. Chloroplast Zetaproteobacteria Rare populations: Alphaproteobacteria Method A log(RNA(%)+1) Zetaproteobacteria log(RNA(%)+1) Method C Method B 0.5 0.5 Cyanobacteria,@ Bacteroidetes Chloroplast Firmicutes Firmicutes Bacteroidetes Planctomycetes Planctomycetes Ac8nobacteria Fig. -
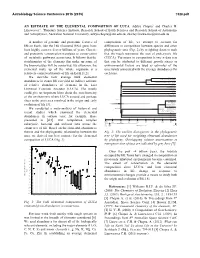
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Establishment and Analysis of Microbial Communities Capable of Producing Methane from Grass Waste at Extremely High C/N Ratio
International Journal of New Technology and Research (IJNTR) ISSN:2454-4116, Volume-2, Issue-9, September 2016 Pages 81-86 Establishment and Analysis of Microbial Communities Capable of Producing Methane from Grass Waste at Extremely High C/N Ratio S. Matsuda, T. Ohtsuki* Anaerobic digestion is a conventional method for biomass Abstract— Acclimation of microbial communities, aiming to utilization [8]. Despite the fact that a great deal of research methane production from grass as a sole substrate at extremely for methane production from grass has been conducted high carbon-to-nitrogen (C/N) ratio, was conducted. In a series especially in recent years, almost processes need supply of of experiments with various sizes of added grass, two microbial communities showing high methane production were obtained abundant sludge source such as sewage and manure [9, 10]. with powdered grass. In the two microbial communities Many studies indicated that the optimal carbon-to-nitrogen designated NR and RP, Bacteroidia including genera (C/N) ratio in methane fermentation were 25-30, and the C/N Bacteroides, Dysgonomonas, Proteiniphilum, and Alistipes were ratio higher than 40 are not generally suitable [11]. The C/N detected as dominant members in eubacteria. It was also shown ration of raw grass shows a wide range; wild grassy weed is at that Methanomicrobia and Methanobacteria including genera relatively high (>59) while lawn grass is at low (<29) [12]. Methanomassiliicoccus and Methanobacterium were found as dominant members in methanogen. It is noteworthy that Therefore, it is difficult to conduct methane fermentation nitrogen fixation were observed both in NR and RP, suggesting with wild grass-only, also being due to less degradability of that insufficiency of nitrogen sources would be complemented lignocellulose [13]. -

Supplementary Information
Retroconversion of estrogens into androgens by bacteria via a cobalamin-mediated methylation Po-Hsiang Wang, Yi-Lung Chen, Sean Ting-Shyang Wei, Kan Wu, Tzong-Huei Lee, Tien-Yu Wu, and Yin-Ru Chiang Supplementary Information Table of Contents Dataset Dataset S1. Genome annotation of strain DHT3 and transcriptomic analysis (RNA-Seq) of bacterial cells grown anaerobically with testosterone or estradiol. SI Tables Table S1. Oligonucleotides used in this study. Table S2. Selection of housekeeping genes of strain DHT3 used for constructing the linear regression line in the global gene expression profiles (RNA-Seq). Table S3. Selection of the cobalamin-dependent methyltransferases used for the un-rooted maximum likelihood tree construction. Table S4. UPLC–APCI–HRMS data of the intermediates involved in anaerobic estrone catabolism by strain DHT3. Table S5. 1H- (600 MHz) and 13C-NMR (150 MHz) spectral data of the HPLC-purified metabolite (AND2) and the authentic standard 5-androstan-3,17-diol Table S6. Selection of the bacteria used for comparative analysis of the gene organization for HIP degradation. SI Figures Fig. S1 Scanning electron micrographs of strain DHT3 cells. Fig. S2 Cobalamin as an essential vitamin during the anaerobic growth of strain DHT3 on estradiol. Fig. S3 Arrangement and expression analysis of the emt genes in strain DHT3. Fig. S4 The anaerobic growth of the wild type (A) and the emtA-disrupted mutant (B) of strain DHT3 with testosterone and estradiol. Fig. S5 APCI–HRMS spectrum of the HIP produced by estrone-fed strain DHT3. 1 Fig. S6 UPLC–APCI–HRMS spectra of two TLC-purified androgen metabolites, 17β-hydroxyandrostan-3-one (A) and 3β,17β-dihydroxyandrostane (B). -

Methanogens Diversity During Anaerobic Sewage Sludge Stabilization and the Effect of Temperature
processes Article Methanogens Diversity during Anaerobic Sewage Sludge Stabilization and the Effect of Temperature Tomáš Vítˇez 1,2, David Novák 3, Jan Lochman 3,* and Monika Vítˇezová 1,* 1 Department of Experimental Biology, Faculty of Science, Masaryk University, 62500 Brno, Czech Republic; [email protected] 2 Department of Agricultural, Food and Environmental Engineering, Faculty of AgriSciences, Mendel University, 61300 Brno, Czech Republic 3 Department of Biochemistry, Faculty of Science, Masaryk University, 62500 Brno, Czech Republic; [email protected] * Correspondence: [email protected] (J.L.); [email protected] (M.V.); Tel.: +420-549-495-602 (J.L.); Tel.: +420-549-497-177 (M.V.) Received: 29 June 2020; Accepted: 10 July 2020; Published: 12 July 2020 Abstract: Anaerobic sludge stabilization is a commonly used technology. Most fermenters are operated at a mesophilic temperature regime. Modern trends in waste management aim to minimize waste generation. One of the strategies can be achieved by anaerobically stabilizing the sludge by raising the temperature. Higher temperatures will allow faster decomposition of organic matter, shortening the retention time, and increasing biogas production. This work is focused on the description of changes in the community of methanogenic microorganisms at different temperatures during the sludge stabilization. At higher temperatures, biogas contained a higher percentage of methane, however, there was an undesirable accumulation of ammonia in the fermenter. Representatives of the hydrogenotrophic genus Methanoliea were described at all temperatures tested. At temperatures up to 50 ◦C, a significant proportion of methanogens were also formed by acetoclastic representatives of Methanosaeta sp. and acetoclastic representatives of the order Methanosarcinales.