Methyl-Coenzyme M Reductase: Expressing Active Recombinant Enzyme in M
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -
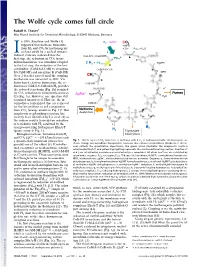
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Xuejun Yu Dissertation Final
UNIVERSITY OF CALIFORNIA RIVERSIDE Conversion of Carbon Dioxide to Formate by a Formate Dehydrogenase from Cupriavidus necator A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Bioengineering by Xuejun Yu September 2018 Dissertation Committee: Dr. Ashok Mulchandani, Co-Chairperson Dr. Xin Ge, Co-Chairperson Dr. Russ Hille Copyright by Xuejun Yu 2018 The Dissertation of Xuejun Yu is approved: Committee Co-Chairperson Committee Co-Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to express sincere appreciation to my advisor, Professor Ashok Mulchandani, for accepting me to be his student when I was suffering. Thank you very much for your delicate guidance, support and encouragement during the past years. You not only guide me on my PhD study, but also help me to be mature on my personality. From bottom of my heart, I feel very lucky to have you as professor. Also, many thanks go to my other committee members. Professor Xin Ge acted as my co-advisor and provided many valuable comments on my molecular cloning work. Professor Russ Hille has also provided insights, encouragements and advices as a member of my committee members. I have learned so many important insights from our meetings and discussions and thank you for bringing me into the molybdenum/tungsten enzyme conference. I am also thankful to the past and current group members, colleagues and friends - Dimitri Niks, Pankaj Ramnani, Feng Tan, Rabeay Hassan, Trupti Terse, Thien-Toan Tran, Claudia Chaves, Jia-wei Tay, Hui Wang, Pham Tung, Hilda Chan, Yingning Gao and Tynan Young. -

192ICM ICBIC Posters
Journal of Inorganic Biochemistry 96 (2003) 203 Monomeric TpPrMoVOSR complexes via the chemical reduction of TpPrMoVIOSR. David J Nielsen, School of Chemistry, University of Melbourne, Australia Christian J Doonan, School of Chemistry, University of Melbourne, Australia Graham N George, Stanford Synchrotron Radiation Laboratory, United States Hugh Harris, Stanford Synchrotron Radiation Laboratory, United States Charles G Young, University of Melbourne, Australia EPR evidence has suggested the presence of molybdenum(V) intermediates in the catalytic cycle of hydroxylase enzyme systems [1], and references therein], and as such these species are attractive targets for the synthesis of small-molecule model systems. Ongoing work in our group has allowed access to several stable and well characterised monomeric molybdenum(VI) oxo-thio complexes TpPrMoVIOSR (TpPr = hydridotris(3-isopropylpyrazol-1-yl)borate) with co-ligand R = eg. substituted phenolates [2], as shown below. These Mo(VI) complexes have proved amenable to chemical reduction using cobaltocene (CoCp2) yielding initially the Pr V [CoCp2][Tp Mo OSR] salts [1,2]. Solution and solid state sulfur X-ray absorption spectroscopy (XAS) on selected examples of the chemically reduced species shows pre-edge features attributable to the S 1s → Mo=S π* transition of a [MoOS]+ unit. Further spectroscopic investigations (EPR, IR) are consistent with the presence of a paramagnetic Mo(V) centre bearing a terminal thio ligand. Continuing spectroscopic, structural and reactivity investigations centred on these important species will be presented. References: [1] P. D. Smith, D. A. Slizys, G. N. George and C. G. Young, J. Amer. Chem. Soc., 122(12), 2000, 2946. [2] C. J. Doonan, Unpublished results. -

Van Heuvelen Department of Chemistry, Harvey Mudd College
Development of Bio-Inspired Catalysts for Dechlorination Reactions Prof. Katherine Van Heuvelen Department of Chemistry, Harvey Mudd College Abstract The nickel-containing cofactor F430 found in methyl-coenzyme M reductase (MCR) and the cobalt-containing cobalamin cofactor (Cbl) found in Vitamin B12 carry out the reductive dehalogenation of chlorinated alkenes, which can act as damaging pollutants in the environment. Both F430 and Cbl are found in biological systems and carry out this reaction under benign conditions using earth-abundant materials. This work centers on the preparation and investigation of small molecular model compounds that reproduce key geometric and electronic features of cofactors F430 and Cbl. In particular, I propose to: 1. Prepare a series of nickel- and cobalt-containing F430 model compounds designed to investigate the influence of the supporting ligand on reactivity. 2. Evaluate the reactivity of these complexes towards halogenated substrates. 3. Characterize reaction intermediates using a combination of spectroscopic (UV-visible, infrared, NMR) and computational (density functional theory, DFT) techniques in order to correlate geometric and electronic structure with reactivity. 4. Elucidate the reaction mechanism using insights gained from aims 1–3, ultimately applying a detailed understanding of the fundamental chemistry underlying dehalogenation to the rational design of an improved catalytic system to treat chlorinated pollutants before they enter the water supply. Start Date, Duration, and Location This research will be conducted at Harvey Mudd College over a ten-week period in the summer of 2016, which will run from May 23 – July 29. The Chemistry Department is in the process of hiring students for the summer of 2016 and the student working on this project will be identified later in the spring semester. -

Deciphering the Mechanism of Enzymatic Methane Synthesis
Science Highlight – June 2021 Deciphering the Mechanism of Enzymatic Methane Synthesis Methane is the simplest organic compound with the highest energy content of any carbon- based fuel. Methane accounts for almost a quarter of U.S. energy consumption, with one-half of homes using natural gas as their heating fuel.1-2 Interestingly, more than 90% of all methane on earth is produced biogenically by methanogens which are responsible for enzymatic synthesis of 1 billion tons of methane per year.3-4 Much of the methane formed by methanogens is captured and used as an energy source by aerobic and anaerobic methanotrophic microbes.5-6 However, the increased mining of methane and industrial farming of cattle has created a mismatch between the sources and the sinks of methane, causing its atmospheric levels to double over the past two centuries. This is an environmental concern related to climate change because pound for pound, methane causes 25 times more 7 global warming over a 100-year period than CO2. Thus, understanding the biosynthesis of methane is imperative from basic energy, economic and environmental perspectives. Methane is formed by one of the few Ni containing cofactors (F430) as part of the active site of methyl coenzyme M reductase (MCR), a strictly anaerobic enzyme present in methanogenic archaea. MCR catalyzes the reaction of methyl-coenzyme M (CH3−SCoM) with coenzyme B (HSCoB) to form methane and the heterodisulfide CoMS−SCoB.8 Besides the importance of methane metabolism in our environment and global energy landscape, the basic chemistry and biology of alkane activation and formation by MCR are of fundamental interest. -

Characterisation, Classification and Conformational Variability Of
Characterisation, Classification and Conformational Variability of Organic Enzyme Cofactors Julia D. Fischer European Bioinformatics Institute Clare Hall College University of Cambridge A thesis submitted for the degree of Doctor of Philosophy 11 April 2011 This dissertation is the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. This dissertation does not exceed the word limit of 60,000 words. Acknowledgements I would like to thank all the members of the Thornton research group for their constant interest in my work, their continuous willingness to answer my academic questions, and for their company during my time at the EBI. This includes Saumya Kumar, Sergio Martinez Cuesta, Matthias Ziehm, Dr. Daniela Wieser, Dr. Xun Li, Dr. Irene Pa- patheodorou, Dr. Pedro Ballester, Dr. Abdullah Kahraman, Dr. Rafael Najmanovich, Dr. Tjaart de Beer, Dr. Syed Asad Rahman, Dr. Nicholas Furnham, Dr. Roman Laskowski and Dr. Gemma Holli- day. Special thanks to Asad for allowing me to use early development versions of his SMSD software and for help and advice with the KEGG API installation, to Roman for knowing where to find all kinds of data, to Dani for help with R scripts, to Nick for letting me use his E.C. tree program, to Tjaart for python advice and especially to Gemma for her constant advice and feedback on my work in all aspects, in particular the chemistry side. Most importantly, I would like to thank Prof. Janet Thornton for giving me the chance to work on this project, for all the time she spent in meetings with me and reading my work, for sharing her seemingly limitless knowledge and enthusiasm about the fascinating world of enzymes, and for being such an experienced and motivational advisor. -

Downloaded from 5Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, United Kingdom
Kent Academic Repository Full text document (pdf) Citation for published version Bryant, Donald A. and Hunter, C. Neil and Warren, Martin J. (2020) Biosynthesis of the modified tetrapyrroles: the pigments of life. Journal of Biological Chemistry . ISSN 0021-9258. DOI https://doi.org/10.1074/jbc.REV120.006194 Link to record in KAR https://kar.kent.ac.uk/80865/ Document Version Author's Accepted Manuscript Copyright & reuse Content in the Kent Academic Repository is made available for research purposes. Unless otherwise stated all content is protected by copyright and in the absence of an open licence (eg Creative Commons), permissions for further reuse of content should be sought from the publisher, author or other copyright holder. Versions of research The version in the Kent Academic Repository may differ from the final published version. Users are advised to check http://kar.kent.ac.uk for the status of the paper. Users should always cite the published version of record. Enquiries For any further enquiries regarding the licence status of this document, please contact: [email protected] If you believe this document infringes copyright then please contact the KAR admin team with the take-down information provided at http://kar.kent.ac.uk/contact.html JBC Papers in Press. Published on April 2, 2020 as Manuscript REV120.006194 The latest version is at https://www.jbc.org/cgi/doi/10.1074/jbc.REV120.006194 Biosynthesis of the modified tetrapyrroles—the pigments of life Donald A. Bryant1,2*, C. Neil Hunter3,‡, and Martin J. Warren4,5,§ 1Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802 2Department of Chemistry and Biochemistry, Montana State University, Bozeman, MT 59717 3Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield S10 2TN, United Kingdom 4School of Biosciences, University of Kent, Canterbury, CT2 7NJ, United Kingdom Downloaded from 5Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, United Kingdom. -

Downloaded from the Genome NCBI
bioRxiv1 preprint doi: https://doi.org/10.1101/765248; this version posted July 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Groundwater Elusimicrobia are metabolically diverse compared to gut microbiome Elusimicrobia 2 and some have a novel nitrogenase paralog. 3 4 Raphaël Méheust1,2, Cindy J. Castelle1,2, Paula B. Matheus Carnevali1,2, Ibrahim F. Farag3, Christine He1, 5 Lin-Xing Chen1,2, Yuki Amano4, Laura A. Hug5, and Jillian F. Banfield1,2,*. 6 7 1Department of Earth and Planetary Science, University of California, Berkeley, Berkeley, CA 94720, 8 USA 9 2Innovative Genomics Institute, Berkeley, CA 94720, USA 10 3School of Marine Science and Policy, University of Delaware, Lewes, DE 19968, USA 11 4Nuclear Fuel Cycle Engineering Laboratories, Japan Atomic Energy Agency, Tokai-mura, Ibaraki, Japan 12 5Department of Biology, University of Waterloo, ON, Canada 13 *Corresponding author: Email: [email protected] 14 15 Abstract 16 Currently described members of Elusimicrobia, a relatively recently defined phylum, are animal- 17 associated and rely on fermentation. However, free-living Elusimicrobia have been detected in sediments, 18 soils and groundwater, raising questions regarding their metabolic capacities and evolutionary 19 relationship to animal-associated species. Here, we analyzed 94 draft-quality, non-redundant genomes, 20 including 30 newly reconstructed genomes, from diverse animal-associated and natural environments. 21 Genomes group into 12 clades, 10 of which previously lacked reference genomes. Groundwater- 22 associated Elusimicrobia are predicted to be capable of heterotrophic or autotrophic lifestyles, reliant on 23 oxygen or nitrate/nitrite-dependent respiration, or a variety of organic compounds and Rhodobacter 24 nitrogen fixation-dependent (Rnf-dependent) acetogenesis with hydrogen and carbon dioxide as the 25 substrates. -

Methane Generation Via Intraprotein C-S Bond Cleavage in Cytochrome
Journal of Organometallic Chemistry 901 (2019) 120945 Contents lists available at ScienceDirect Journal of Organometallic Chemistry journal homepage: www.elsevier.com/locate/jorganchem Methane generation via intraprotein CeS bond cleavage in cytochrome b562 reconstituted with nickel didehydrocorrin * ** Yuta Miyazaki a, Koji Oohora a, b, , Takashi Hayashi a, a Department of Applied Chemistry, Graduate School of Engineering, Osaka University, Suita, 565-0871, Japan b Frontier Research Base for Global Young Researchers, Graduate School of Engineering, Osaka University, Suita, 565-0871, Japan article info abstract II Article history: Cytochrome b562 (Cyt b562) reconstituted with nickel didehydrocorrin (Ni (DDHC)), a protein-based Received 20 August 2019 functional model of methyl-coenzyme M reductase (MCR), was investigated to demonstrate methane Received in revised form generation via intraprotein cleavage of a CeS bond. NiII(DDHC) was synthesized as a model complex of an 16 September 2019 MCR cofactor known as F430 and found to show NiII/NiI redox behavior with a potential of À0.61 V vs. Accepted 18 September 2019 AgjAgCl. This potential is slightly positive-shifted compared to that of F430 without protein. Conjugation Available online 19 September 2019 II II of Ni (DDHC) with the apo-form of Cyt b562 provides reconstituted Cyt b562 (rCyt b562(Ni (DDHC))) which II was characterized by spectroscopic measurements. Photoirradiation of rCyt b562(Ni (DDHC)) generates Keywords: 0 Hemoprotein methane gas in the presence of tris(2,2 -bipyridine)ruthenium(II) chloride as a photosensitizer and so- fi MCR dium ascorbate as a sacri cial reagent. Further experiments using Cyt b562 mutants indicate that F430 methane is derived from the CH3S group of the methionine residue in the heme-binding site where Methane thioether, thiol and the nickel center are precisely arranged. -

A Niii-Containing Vitamin B12 Derivative with a Cofactor-F430
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2018 A Nickel(II)-Containing Vitamin B12 Derivative with a Cofactor-F430-type -System Brenig, Christopher ; Prieto, Lucas ; Oetterli, René ; Zelder, Felix Abstract: F430 is a unique enzymatic cofactor in the production and oxidation of methane by strictly anaerobic bacteria. The key enzyme methyl coenzyme M reductase (MCR) contains a hydroporphi- noid nickel complex with a characteristic absorption maximum at around 430 nm in its active site. Herein, the three‐step semisynthesis of a hybrid NiII‐containing corrinoid that partly resembles F430 in its structural and spectroscopic features from vitamin B12 is presented. A key step of the route is the simultaneous demetalation and ring closure reaction of a 5,6‐secocobalamin to metal‐free 5,6‐dihy- droxy‐5,6‐dihydrohydrogenobalamin with cobaltocene and KCN under reductive conditions. Studies on the coordination chemistry of the novel compound support an earlier hypothesis why nature carefully selected a corphin over a corrin ligand in F430 for challenging nickel‐catalyzed biochemical reactions. DOI: https://doi.org/10.1002/anie.201810983 Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-183474 Journal Article Accepted Version Originally published at: Brenig, Christopher; Prieto, Lucas; Oetterli, René; Zelder, Felix (2018). A Nickel(II)-Containing Vita- min B12 Derivative with a Cofactor-F430-type -System. Angewandte Chemie Internationale Edition, 57(50):16308-16312. DOI: https://doi.org/10.1002/anie.201810983 II A Ni -Containing Vitamin B12 Derivative with a Cofactor-F430-Type π-System Christopher Brenig‡, Lucas Prieto‡, René Oetterli and Felix Zelder* Department of Chemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland. -

Methylotrophic Methanogenesis in Hydraulically Fractured Shales
Methylotrophic Methanogenesis in Hydraulically Fractured Shales Master’s Thesis Presented in Partial Fulfillment of the Requirements for the Degree Master of Science in the Graduate School of The Ohio State University By: Daniel Nimmer Marcus, B.S. Graduate Program in Microbiology The Ohio State University 2016 Thesis Committee: Kelly Wrighton PhD, Advisor Joseph Krzycki PhD Charles Daniels PhD Matthew Sullivan PhD Copyright by Daniel Nimmer Marcus 2016 ABSTRACT Over the last decade shale gas obtained from hydraulic fracturing of deep shale formations has become a sizeable component of the US energy portfolio. There is a growing body of evidence indicating that methanogenic archaea are both present and active in hydraulically fractured shales. However, little is known about the genomic architecture of shale derived methanogens. Here we leveraged natural gas extraction activities in the Appalachian region to gain access to fluid samples from two geographically and geologically distinct shale formations. Samples were collected over a time series from both shales for a period of greater than eleven months. Using assembly based metagenomics, two methanogen genomes from the genus Methanohalophilus were recovered and estimated to be near complete (97.1 and 100%) by 104 archaeal single copy genes. Additionally, a Methanohalophilus isolate was obtained which yielded a genome estimated to be 100% complete by the same metric. Based on metabolic reconstruction, it is inferred that these organisms utilize C-1 methyl substrates for methanogenesis. The ability to utilize monomethylamine, dimethylamine and methanol was experimentally confirmed with the Methanohalophilus isolate. In situ concentrations of C-1 methyl substrates, osmoprotectants, and Cl- were measured in parallel with estimates of community membership.