The Wolfe Cycle Comes Full Circle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Methanothermus Fervidus Type Strain (V24S)
UC Davis UC Davis Previously Published Works Title Complete genome sequence of Methanothermus fervidus type strain (V24S). Permalink https://escholarship.org/uc/item/9367m39j Journal Standards in genomic sciences, 3(3) ISSN 1944-3277 Authors Anderson, Iain Djao, Olivier Duplex Ngatchou Misra, Monica et al. Publication Date 2010-11-20 DOI 10.4056/sigs.1283367 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Standards in Genomic Sciences (2010) 3:315-324 DOI:10.4056/sigs.1283367 Complete genome sequence of Methanothermus fervidus type strain (V24ST) Iain Anderson1, Olivier Duplex Ngatchou Djao2, Monica Misra1,3, Olga Chertkov1,3, Matt Nolan1, Susan Lucas1, Alla Lapidus1, Tijana Glavina Del Rio1, Hope Tice1, Jan-Fang Cheng1, Roxanne Tapia1,3, Cliff Han1,3, Lynne Goodwin1,3, Sam Pitluck1, Konstantinos Liolios1, Natalia Ivanova1, Konstantinos Mavromatis1, Natalia Mikhailova1, Amrita Pati1, Evelyne Brambilla4, Amy Chen5, Krishna Palaniappan5, Miriam Land1,6, Loren Hauser1,6, Yun-Juan Chang1,6, Cynthia D. Jeffries1,6, Johannes Sikorski4, Stefan Spring4, Manfred Rohde2, Konrad Eichinger7, Harald Huber7, Reinhard Wirth7, Markus Göker4, John C. Detter1, Tanja Woyke1, James Bristow1, Jonathan A. Eisen1,8, Victor Markowitz5, Philip Hugenholtz1, Hans-Peter Klenk4, and Nikos C. Kyrpides1* 1 DOE Joint Genome Institute, Walnut Creek, California, USA 2 HZI – Helmholtz Centre for Infection Research, Braunschweig, Germany 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA 4 DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany 5 Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, California, USA 6 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA 7 University of Regensburg, Archaeenzentrum, Regensburg, Germany 8 University of California Davis Genome Center, Davis, California, USA *Corresponding author: Nikos C. -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

Histone Variants in Archaea and the Evolution of Combinatorial Chromatin Complexity
Histone variants in archaea and the evolution of combinatorial chromatin complexity Kathryn M. Stevensa,b, Jacob B. Swadlinga,b, Antoine Hochera,b, Corinna Bangc,d, Simonetta Gribaldoe, Ruth A. Schmitzc, and Tobias Warneckea,b,1 aMolecular Systems Group, Quantitative Biology Section, Medical Research Council London Institute of Medical Sciences, London W12 0NN, United Kingdom; bInstitute of Clinical Sciences, Faculty of Medicine, Imperial College London, London W12 0NN, United Kingdom; cInstitute for General Microbiology, University of Kiel, 24118 Kiel, Germany; dInstitute of Clinical Molecular Biology, University of Kiel, 24105 Kiel, Germany; and eDepartment of Microbiology, Unit “Evolutionary Biology of the Microbial Cell,” Institut Pasteur, 75015 Paris, France Edited by W. Ford Doolittle, Dalhousie University, Halifax, NS, Canada, and approved October 28, 2020 (received for review April 14, 2020) Nucleosomes in eukaryotes act as platforms for the dynamic inte- additional histone dimers can be taggedontothistetramertoyield gration of epigenetic information. Posttranslational modifications oligomers of increasing length that wrap correspondingly more DNA are reversibly added or removed and core histones exchanged for (3, 6–9). Almost all archaeal histones lack tails and PTMs have yet to paralogous variants, in concert with changing demands on tran- be reported. Many archaea do, however, encode multiple histone scription and genome accessibility. Histones are also common in paralogs (8, 10) that can flexibly homo- and heterodimerize in -
The Wolfe Cycle Comes Full Circle
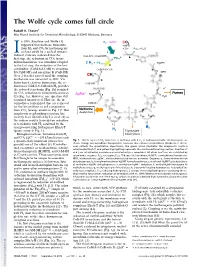
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Supporting Information
Supporting Information Lozupone et al. 10.1073/pnas.0807339105 SI Methods nococcus, and Eubacterium grouped with members of other Determining the Environmental Distribution of Sequenced Genomes. named genera with high bootstrap support (Fig. 1A). One To obtain information on the lifestyle of the isolate and its reported member of the Bacteroidetes (Bacteroides capillosus) source, we looked at descriptive information from NCBI grouped firmly within the Firmicutes. This taxonomic error was (www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and other related not surprising because gut isolates have often been classified as publications. We also determined which 16S rRNA-based envi- Bacteroides based on an obligate anaerobe, Gram-negative, ronmental surveys of microbial assemblages deposited near- nonsporulating phenotype alone (6, 7). A more recent 16S identical sequences in GenBank. We first downloaded the gbenv rRNA-based analysis of the genus Clostridium defined phylo- files from the NCBI ftp site on December 31, 2007, and used genetically related clusters (4, 5), and these designations were them to create a BLAST database. These files contain GenBank supported in our phylogenetic analysis of the Clostridium species in the HGMI pipeline. We thus designated these Clostridium records for the ENV database, a component of the nonredun- species, along with the species from other named genera that dant nucleotide database (nt) where 16S rRNA environmental cluster with them in bootstrap supported nodes, as being within survey data are deposited. GenBank records for hits with Ͼ98% these clusters. sequence identity over 400 bp to the 16S rRNA sequence of each of the 67 genomes were parsed to get a list of study titles Annotation of GTs and GHs. -

Evaluation of Methanobrevibacter Smithii As a Human-Specific Marker
The University of Southern Mississippi The Aquila Digital Community Presentations Microbial Source Tracking 2005 Evaluation of Methanobrevibacter smithii as a Human-Specific aM rker of Fecal Pollution Jennifer A. Ufnar University of Southern Mississippi Shiao Y. Wang University of Southern Mississippi R.D. Ellender University of Southern Mississippi Follow this and additional works at: https://aquila.usm.edu/mst_presentations Recommended Citation Ufnar, Jennifer A.; Wang, Shiao Y.; and Ellender, R.D., "Evaluation of Methanobrevibacter smithii as a Human-Specific aM rker of Fecal Pollution" (2005). Presentations. 5. https://aquila.usm.edu/mst_presentations/5 This Poster is brought to you for free and open access by the Microbial Source Tracking at The Aquila Digital Community. It has been accepted for inclusion in Presentations by an authorized administrator of The Aquila Digital Community. For more information, please contact [email protected]. Evaluation of Methanobrevibacter smithii as a Human-Specific Q-311 For More Information Contact: Marker of Fecal Pollution Dr. R.D. Ellender University of Southern Mississippi Department of Biological Sciences Jennifer A. Ufnar, Shiao Y. Wang, and R.D. Ellender 118 College Drive #5018 Hattiesburg, MS 39406-0001 University of Southern Mississippi, Hattiesburg, MS 601-266-4720 or 601-266-4752(lab) Results Abstract Primers specific for the Methanobrevibacter genus (MET) amplified a product of 282bp. The Mnif primers amplified a 222bp product. The MET primers were Microbial source tracking has historically focused on the origin of traditional enteric indicators including coliforms, enterococci, or Escherichia coli. Recently, positive for all Methanobrevibacter species, and did not amplify a product in any other bacterial or methanogen genus. -

Xuejun Yu Dissertation Final
UNIVERSITY OF CALIFORNIA RIVERSIDE Conversion of Carbon Dioxide to Formate by a Formate Dehydrogenase from Cupriavidus necator A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Bioengineering by Xuejun Yu September 2018 Dissertation Committee: Dr. Ashok Mulchandani, Co-Chairperson Dr. Xin Ge, Co-Chairperson Dr. Russ Hille Copyright by Xuejun Yu 2018 The Dissertation of Xuejun Yu is approved: Committee Co-Chairperson Committee Co-Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to express sincere appreciation to my advisor, Professor Ashok Mulchandani, for accepting me to be his student when I was suffering. Thank you very much for your delicate guidance, support and encouragement during the past years. You not only guide me on my PhD study, but also help me to be mature on my personality. From bottom of my heart, I feel very lucky to have you as professor. Also, many thanks go to my other committee members. Professor Xin Ge acted as my co-advisor and provided many valuable comments on my molecular cloning work. Professor Russ Hille has also provided insights, encouragements and advices as a member of my committee members. I have learned so many important insights from our meetings and discussions and thank you for bringing me into the molybdenum/tungsten enzyme conference. I am also thankful to the past and current group members, colleagues and friends - Dimitri Niks, Pankaj Ramnani, Feng Tan, Rabeay Hassan, Trupti Terse, Thien-Toan Tran, Claudia Chaves, Jia-wei Tay, Hui Wang, Pham Tung, Hilda Chan, Yingning Gao and Tynan Young. -

Archaea: Essential Inhabitants of the Human Digestive Microbiota Vanessa Demonfort Nkamga, Bernard Henrissat, Michel Drancourt
Archaea: Essential inhabitants of the human digestive microbiota Vanessa Demonfort Nkamga, Bernard Henrissat, Michel Drancourt To cite this version: Vanessa Demonfort Nkamga, Bernard Henrissat, Michel Drancourt. Archaea: Essential inhabi- tants of the human digestive microbiota. Human Microbiome Journal, Elsevier, 2017, 3, pp.1-8. 10.1016/j.humic.2016.11.005. hal-01803296 HAL Id: hal-01803296 https://hal.archives-ouvertes.fr/hal-01803296 Submitted on 8 Jun 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Human Microbiome Journal 3 (2017) 1–8 Contents lists available at ScienceDirect Human Microbiome Journal journal homepage: www.elsevier.com/locate/humic Archaea: Essential inhabitants of the human digestive microbiota ⇑ Vanessa Demonfort Nkamga a, Bernard Henrissat b, Michel Drancourt a, a Aix Marseille Univ, INSERM, CNRS, IRD, URMITE, Marseille, France b Aix-Marseille-Université, AFMB UMR 7257, Laboratoire «architecture et fonction des macromolécules biologiques», Marseille, France article info abstract Article history: Prokaryotes forming the -

4 Metabolic and Taxonomic Diversification in Continental Magmatic Hydrothermal Systems
Maximiliano J. Amenabar, Matthew R. Urschel, and Eric S. Boyd 4 Metabolic and taxonomic diversification in continental magmatic hydrothermal systems 4.1 Introduction Hydrothermal systems integrate geological processes from the deep crust to the Earth’s surface yielding an extensive array of spring types with an extraordinary diversity of geochemical compositions. Such geochemical diversity selects for unique metabolic properties expressed through novel enzymes and functional characteristics that are tailored to the specific conditions of their local environment. This dynamic interaction between geochemical variation and biology has played out over evolu- tionary time to engender tightly coupled and efficient biogeochemical cycles. The timescales by which these evolutionary events took place, however, are typically in- accessible for direct observation. This inaccessibility impedes experimentation aimed at understanding the causative principles of linked biological and geological change unless alternative approaches are used. A successful approach that is commonly used in geological studies involves comparative analysis of spatial variations to test ideas about temporal changes that occur over inaccessible (i.e. geological) timescales. The same approach can be used to examine the links between biology and environment with the aim of reconstructing the sequence of evolutionary events that resulted in the diversity of organisms that inhabit modern day hydrothermal environments and the mechanisms by which this sequence of events occurred. By combining molecu- lar biological and geochemical analyses with robust phylogenetic frameworks using approaches commonly referred to as phylogenetic ecology [1, 2], it is now possible to take advantage of variation within the present – the distribution of biodiversity and metabolic strategies across geochemical gradients – to recognize the extent of diversity and the reasons that it exists. -

Variations in the Two Last Steps of the Purine Biosynthetic Pathway in Prokaryotes
GBE Different Ways of Doing the Same: Variations in the Two Last Steps of the Purine Biosynthetic Pathway in Prokaryotes Dennifier Costa Brandao~ Cruz1, Lenon Lima Santana1, Alexandre Siqueira Guedes2, Jorge Teodoro de Souza3,*, and Phellippe Arthur Santos Marbach1,* 1CCAAB, Biological Sciences, Recoˆ ncavo da Bahia Federal University, Cruz das Almas, Bahia, Brazil 2Agronomy School, Federal University of Goias, Goiania,^ Goias, Brazil 3 Department of Phytopathology, Federal University of Lavras, Minas Gerais, Brazil Downloaded from https://academic.oup.com/gbe/article/11/4/1235/5345563 by guest on 27 September 2021 *Corresponding authors: E-mails: [email protected]fla.br; [email protected]. Accepted: February 16, 2019 Abstract The last two steps of the purine biosynthetic pathway may be catalyzed by different enzymes in prokaryotes. The genes that encode these enzymes include homologs of purH, purP, purO and those encoding the AICARFT and IMPCH domains of PurH, here named purV and purJ, respectively. In Bacteria, these reactions are mainly catalyzed by the domains AICARFT and IMPCH of PurH. In Archaea, these reactions may be carried out by PurH and also by PurP and PurO, both considered signatures of this domain and analogous to the AICARFT and IMPCH domains of PurH, respectively. These genes were searched for in 1,403 completely sequenced prokaryotic genomes publicly available. Our analyses revealed taxonomic patterns for the distribution of these genes and anticorrelations in their occurrence. The analyses of bacterial genomes revealed the existence of genes coding for PurV, PurJ, and PurO, which may no longer be considered signatures of the domain Archaea. Although highly divergent, the PurOs of Archaea and Bacteria show a high level of conservation in the amino acids of the active sites of the protein, allowing us to infer that these enzymes are analogs. -

Tungsten-Enhanced Growth of Methanosphaera Stadtmanae Bédis Dridi1, Saber Khelaifia1, Marie-Laure Fardeau2, Bernard Ollivier2 and Michel Drancourt1*
Dridi et al. BMC Research Notes 2012, 5:238 http://www.biomedcentral.com/1756-0500/5/238 SHORT REPORT Open Access Tungsten-enhanced growth of Methanosphaera stadtmanae Bédis Dridi1, Saber Khelaifia1, Marie-Laure Fardeau2, Bernard Ollivier2 and Michel Drancourt1* Abstract Background: The methanogenic Archaea Methanosphaera stadtmanae has been detected in the human gut microbiota by both culture and culture-independent methods. Its growth reaches an exponential phase after 5 to 7-day culture in medium 322 (10% vol). Our recent successful isolation of Methanomassiliicoccus luminyensis,a tungstate-selenite-requiring Archaea sharing similar metabolism characteristics with M. stadtmanae prompted us to study the effects of tungsten and selenium on M. stadtmanae growth. Findings: Addition of 0.2 mg/L sodium tungstate to medium 322 yielded, 48 hours after inoculation, a growth rate equivalent to that obtained after 6 days with control culture as measured by methane monitoring and optical density measurement. Addition of 50 μg/mL sodium selenate had no effect on M. stadtmanae growth. Quantitative real-time PCRs targeting the M. stadtmanae 16S rRNA confirmed these data. Conclusions: These data provide new information regarding the poorly known nutritional requirements of the human gut colonizing organisms M. stadtmanae. Adding sodium tungstate to basal medium may facilitate phenotypic characterization of this organism and additionally aid the isolation of new Archaea from complex host microbiota. Keywords: Methanogenic Archaea, Methanosphaera stadtmanae, Methanomassiliicoccus luminyensis, Tungsten, Selenium Findings [5,6]) within the order Methanobacteriales. We recently Methanosphaera stadtmanae is a spherical-shaped, non- isolated Methanomassiliicoccus luminyensis,thefirstcul- motile archaeon initially isolated from human feces [1]. tured representative of new order of methanoarchaea [7]. -

Taxonomy and Ecology of Methanogens
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Horizon / Pleins textes FEMS Microbiology Reviews 87 (1990) 297-308 297 Pubfished by Elsevier FEMSRE 00180 Taxonomy and ecology of methanogens J.L. Garcia Laboratoire de Microbiologie ORSTOM, Université de Provence, Marseille, France Key words: Methanogens; Archaebacteria; Taxonomy; Ecology 1. INTRODUCTION methane from CO2 using alcohols as hydrogen donors; 2-propanol is oxidized to acetone, and More fhan nine reviews on taxonomy of 2-butanol to 2-butanone. Carbon monoxide may methanogens have been published during the last also be converted into methane; most hydro- decade [l-91, after the discovery of the unique genotrophic species (60%) will also use formate. biochemical and genetic properties of these Some aceticlastic species are incapable of oxidiz- organisms led to the concept of Archaebacteria at ing H,. The aceticlastic species of the genus the end of the seventies. Moreover, important Methanosurcina are the most metabolically diverse economic factors have ,placed these bacteria in the methanogens, whereas the obligate aceticlastic limelight [5], including the need to develop alter- Methanosaeta (Methanothrix) can use only acetate. native forms of energy, xenobiotic pollution con- The taxonomy of the methanogenic bacteria trol, the enhancement of meat yields in the cattle has been extensively revised in the light of new industry, the distinction between biological and information based on comparative studies of 16 S thermocatalytic petroleum generation, and the rRNA oligonucleotide sequences, membrane lipid global distribution of methane in the earth's atmo- composition, and antigenic fingerprinting data. sphere. The phenotypic characteristics often do not pro- vide a sufficient means of distinguishing among taxa or determining the phylogenetic position of a 2.