Generate Metabolic Map Poster
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Heterologous Expression and Identification of the Genes Involved
Heterologous Expression and Identification of the Genes Involved in Anaerobic Degradation of 1,3-Dihydroxybenzene (Resorcinol) in Azoarcus anaerobiusᰔ Paula I. Darley,1† Jutta A. Hellstern,1†‡ Javier I. Medina-Bellver,2 Silvia Marque´s,2 Bernhard Schink,1 and Bodo Philipp1* Fachbereich Biologie, Universita¨t Konstanz, D-78457 Constance, Germany,1 and Estacio´n Experimental del Zaidı´n, C/. Profesor Albareda 1, E-18008 Granada, Spain2 Received 9 November 2006/Accepted 9 March 2007 Azoarcus anaerobius, a strictly anaerobic, gram-negative bacterium, utilizes resorcinol as a sole carbon and energy source with nitrate as an electron acceptor. Previously, we showed that resorcinol degradation by this bacterium is initiated by two oxidative steps, both catalyzed by membrane-associated enzymes that lead to the formation of hydroxyhydroquinone (HHQ; 1,2,4-benzenetriol) and 2-hydroxy-1,4-benzoquinone (HBQ). This study presents evidence for the further degradation of HBQ in cell extracts to form acetic and malic acids. To identify the A. anaerobius genes required for anaerobic resorcinol catabolism, a cosmid library with genomic DNA was constructed and transformed into the phylogenetically related species Thauera aromatica, which cannot grow with resorcinol. By heterologous complementation, a transconjugant was identified that gained the ability to metabolize resorcinol. Its cosmid, designated R؉, carries a 29.88-kb chromosomal DNA fragment containing 22 putative genes. In cell extracts of T. aromatica transconjugants, resorcinol was degraded to HHQ, HBQ, and acetate, suggesting that cosmid R؉ carried all of the genes necessary for resorcinol degradation. On the basis of the physiological characterization of T. aromatica transconjugants carrying transposon insertions -in different genes of cosmid R؉, eight open reading frames were found to be essential for resorcinol miner alization. -

Metaproteogenomic Insights Beyond Bacterial Response to Naphthalene
ORIGINAL ARTICLE ISME Journal – Original article Metaproteogenomic insights beyond bacterial response to 5 naphthalene exposure and bio-stimulation María-Eugenia Guazzaroni, Florian-Alexander Herbst, Iván Lores, Javier Tamames, Ana Isabel Peláez, Nieves López-Cortés, María Alcaide, Mercedes V. del Pozo, José María Vieites, Martin von Bergen, José Luis R. Gallego, Rafael Bargiela, Arantxa López-López, Dietmar H. Pieper, Ramón Rosselló-Móra, Jesús Sánchez, Jana Seifert and Manuel Ferrer 10 Supporting Online Material includes Text (Supporting Materials and Methods) Tables S1 to S9 Figures S1 to S7 1 SUPPORTING TEXT Supporting Materials and Methods Soil characterisation Soil pH was measured in a suspension of soil and water (1:2.5) with a glass electrode, and 5 electrical conductivity was measured in the same extract (diluted 1:5). Primary soil characteristics were determined using standard techniques, such as dichromate oxidation (organic matter content), the Kjeldahl method (nitrogen content), the Olsen method (phosphorus content) and a Bernard calcimeter (carbonate content). The Bouyoucos Densimetry method was used to establish textural data. Exchangeable cations (Ca, Mg, K and 10 Na) extracted with 1 M NH 4Cl and exchangeable aluminium extracted with 1 M KCl were determined using atomic absorption/emission spectrophotometry with an AA200 PerkinElmer analyser. The effective cation exchange capacity (ECEC) was calculated as the sum of the values of the last two measurements (sum of the exchangeable cations and the exchangeable Al). Analyses were performed immediately after sampling. 15 Hydrocarbon analysis Extraction (5 g of sample N and Nbs) was performed with dichloromethane:acetone (1:1) using a Soxtherm extraction apparatus (Gerhardt GmbH & Co. -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

Compression of Large Sets of Sequence Data Reveals Fine Diversification of Functional Profiles in Multigene Families of Proteins
Technical note Compression of Large Sets of Sequence Data Reveals Fine Diversification of Functional Profiles in Multigene Families of Proteins: A Study for Peptidyl-Prolyl cis/trans Isomerases (PPIase) Andrzej Galat Retired from: Service d’Ingénierie Moléculaire des Protéines (SIMOPRO), CEA-Université Paris-Saclay, France; [email protected]; Tel.: +33-0164465072 Received: 21 December 2018; Accepted: 21 January 2019; Published: 11 February 2019 Abstract: In this technical note, we describe analyses of more than 15,000 sequences of FK506- binding proteins (FKBP) and cyclophilins, also known as peptidyl-prolyl cis/trans isomerases (PPIases). We have developed a novel way of displaying relative changes of amino acid (AA)- residues at a given sequence position by using heat-maps. This type of representation allows simultaneous estimation of conservation level in a given sequence position in the entire group of functionally-related paralogues (multigene family of proteins). We have also proposed that at least two FKBPs, namely FKBP36, encoded by the Fkbp6 gene and FKBP51, encoded by the Fkbp5 gene, can form dimers bound via a disulfide bridge in the nucleus. This type of dimer may have some crucial function in the regulation of some nuclear complexes at different stages of the cell cycle. Keywords: FKBP; cyclophilin; PPIase; heat-map; immunophilin 1 Introduction About 30 years ago, an exciting adventure began in finding some correlations between pharmacological activities of macrocyclic hydrophobic drugs, namely the cyclic peptide cyclosporine A (CsA), and two macrolides, namely FK506 and rapamycin, which have profound and clinically useful immunosuppressive effects, especially in organ transplantations and in combating some immune disorders. -

Table S1-Final.Xlsx
Table S1. Functional gene families covered on the GeoChip 5.0. No. of sequence‐ No. of group‐ No. of total No. of covered No. of total Category Subcategory Subcategory 2 Gene Encoded Enzyme specific probesa specific probesa probes on 5M CDSa probes on 5S Categories for Geochemical Cycling Carbon Cycling Carbon Degradation Agar beta_agarase Agarase 42 79 217 121 Agar Total 42 79 0 217 121 Alginate alginase Alginase 45 200 512 245 Alginate Total 45 200 0 512 245 Cellulose Endoglucanase Endoglucanase 198 248 446 740 343 Cellulose Total 198 248 446 740 343 Chitin Acetylglucosaminidase Acetylglucosaminidase 119 976 1095 3088 1116 Chitinase Chitinase 649 1231 1880 3285 1411 endochitinase Endochitinase 213 334 1104 547 exochitinase Exochitinase 14 55 259 69 Chitin Total 995 2596 2975 7736 3143 Glyoxylate cycle AceA Isocitrate lyase 69 373 442 887 0 AceB Malate synthase A 90 610 700 1457 0 Glyoxylate cycle Total 159 983 1142 2344 0 Hemicellulose Ara Arabinofuranosidase 172 543 715 1367 827 Mannanase Mannanase 159 238 397 639 478 Xylanase Xylanase 150 653 803 1739 858 Hemicellulose Total 481 1434 1915 3745 2163 Heparin heparinase Heparinase 7 53 166 60 Heparin Total 7 53 0 166 60 Hyaluronic acid hyaluronidase Hyaluronidase 880 383 88 Hyaluronidase Total 8 80 0 383 88 Lignin Glx Glyoxal oxidase 103 40 143 178 125 Mnp Manganese peroxidase 50 13 63 74 66 Phenol_oxidase Laccase or phenol oxidase 188 372 560 1009 677 Lignin Total 341 425 766 1261 868 Pectin Pectinase (pectate_lyase) Pectate lyase 53 252 305 827 321 Pme Pectin methylesterase 25 240 265 -
The Wolfe Cycle Comes Full Circle
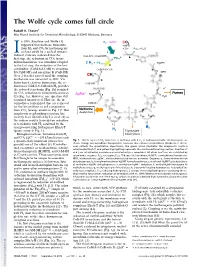
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Synthesis and Application of Light-Switchable Arylazopyrazole Rapamycin Analogs
Organic & Biomolecular Chemistry Synthesis and Application of Light-Switchable Arylazopyrazole Rapamycin Analogs Journal: Organic & Biomolecular Chemistry Manuscript ID OB-COM-08-2019-001719.R1 Article Type: Communication Date Submitted by the 25-Aug-2019 Author: Complete List of Authors: Courtney, Taylor; University of Pittsburgh, Chemistry Horst, Trevor; University of Pittsburgh, Chemistry Hankinson, Chasity; University of Pittsburgh, Chemistry Deiters, Alexander; University of Pittsburgh, Chemistry Page 1 of 11 Organic & Biomolecular Chemistry Synthesis and Application of Light-Switchable Arylazopyrazole Rapamycin Analogs Taylor M. Courtney, Trevor J. Horst, Chasity P. Hankinson, and Alexander Deiters* Department of Chemistry, University of Pittsburgh, Pittsburgh, PA 15260, United States [email protected] Abstract: Rapamycin-induced dimerization of FKBP and FRB has been utilized as a tool for co-localizing two proteins of interest in numerous applications. Due to the tight binding interaction of rapamycin with FKBP and FRB, the ternary complex formation is essentially irreversible. Since biological processes occur in a highly dynamic fashion with cycles of protein association and dissociation to generate a cellular response, it is useful to have chemical tools that function in a similar manner. We have developed arylazopyrazole-modified rapamycin analogs which undergo a configurational change upon light exposure and we observed enhanced ternary complex formation for the cis-isomer over the trans-isomer for one of the analogs. Introduction: Chemical inducers of dimerization (CIDs) are prominent tools used by chemical biologists to place biological processes under conditional control.1-4 The most commonly utilized CID is rapamycin, a natural product that binds to FK506 binding protein (FKBP) with a 0.2 nM Kd. -

The Immunophilins, Fk506 Binding Protein and Cyclophilin, Are Discretely Localized in the Brain: Relationship to Calcineurin
NeuroscienceVol. 62,NO. 2, pp. 569-580,1994 Elsevier Sctence Ltd Copyright 0 1994 IBRO Pergamon 0306-4522(94)E0182-4 Printed in Great Britain. All rights reserved 0306-4522194 $7.00 + 0.00 THE IMMUNOPHILINS, FK506 BINDING PROTEIN AND CYCLOPHILIN, ARE DISCRETELY LOCALIZED IN THE BRAIN: RELATIONSHIP TO CALCINEURIN T. M. DAWSON,*t J. P. STEINER,* W. E. LYONS,*11 M. FOTUHI,* M. BLUE? and S. H. SNYDER*f§l Departments of *Neuroscience, tNeurology, $Pharmacology and Molecular Sciences, and §Psychiatry, Johns Hopkins University School of Medicine, 725 N. Wolfe Street, Baltimore, MD 21205, U.S.A. (IDivision of Toxicological Science, Johns Hopkins University School of Hygiene and Public Health Abstract-The immunosuppressant drugs cyclosporin A and FK506 bind to small, predominantly soluble proteins cyclophilin and FK506 binding protein, respectively, to mediate their pharmacological actions. The immunosuppressant actions of these drugs occur through binding of cyclophilin-cyclosporinA and FK506 binding protein-FK506 complexes to the calcium-calmodulin-dependent protein phosphatase, calcineurin, inhibiting phosphatase activity, Utilizing immunohistcchemistry, in situ hybridization and autoradiography, we have localized protein and messenger RNA for FKS06 binding protein, cyclophilin and calcineurin. All three proteins and/or messages exhibit a heterogenous distribution through the brain and spinal cord, with the majority of the localizations being neuronal. We observe a striking co-localiz- ation of FK506 binding protein and calcineurin in most -

Tannin Degradation by Phytopathogen's Tannase: a Plant's
Biocatalysis and Agricultural Biotechnology 21 (2019) 101342 Contents lists available at ScienceDirect Biocatalysis and Agricultural Biotechnology journal homepage: http://www.elsevier.com/locate/bab Tannin degradation by phytopathogen’s tannase: A Plant’s defense perspective Kanti Prakash Sharma Department of Biosciences, Mody University of Science and Technology, Lakshmangarh, Sikar, Rajasthan, 332311, India ARTICLE INFO ABSTRACT Keywords: Tannins are plant secondary metabolites and characterized as plant defensive molecules. They impose a barrier Tannin against phytopathogens invasion in plants and thus oppose diseases occurrence in them. Tannase is an enzyme Tannase known for its ability to degrade plant tannins. Important plant pathogens also possess tannase coding sequence in Plant defense their genome. Researches on tannase are till date focused on its applications in animal nutrition, in bioreme Pathogen diation and in food industries etc. The information about tannase role with respect to pathogen’s virulence is Virulence very scanty or almost nil in scientific literature. The presence of tannase in pathogen’s genome is an adaptive feature which may be a part of its strategy to overcome the negative effects of plants tannins. The present review summarizes important aspects of tannase and its possible role in disease causing ability of pathogens in plants. 1. Introduction hydrolyzable tannins (Zhang et al., 2001). In complex tannins, a cate chin or epicatechin unit is bound glycosidically to a gallotannin or an Tannins are universally present in plants and represent the fourth ellagitannin unit to yield catechin or epicatechin and gallic acid or most abundant group of secondary metabolites after cellulose, hemi ellagic acid upon hydrolysis. -

Maleylacetate Reductase from Trichosporon Cutaneum
Biochem. J. (1980) 185, 783-786 783 Printed in Great Britain Maleylacetate Reductase from Trichosporon cutaneum Andras B. GAAL and Halina Y. NEUJAHR Department ofBiochemistry and Biochemical Technology, The Royal Institute of Technology, S 100 44 Stockholm, Sweden (Received 12 November 1979) The enzyme catalysing the reduction of maleylacetate to 3-oxoadipate was purified 150-fold from Trichosporon cutaneum, induced for aromatic metabolism by growth with resorcinol as a major carbon source. The enzyme separated upon electrofocusing into three species with pI values 4.6, 5.1 and 5.6. They had similar catalytic properties and the same molecular weight. Our group has previously reported that the yeast fer, pH 7.6, 0.2pmol of maleylacetate, 0.2,mol of Trichosporon cutaneum can be induced to metabol- NADPH and 50-200#ug of enzyme protein. Under ize phenol and resorcinol, both phenols being these conditions 1 enzyme unit corresponds to an attacked by the same hydroxylating enzyme and the absorbance decrease of 6.1 A340 units/min. same ring-cleavage enzyme (Neujahr & Varga, Determination of molecular weight. Molecular 1970; Varga & Neujahr, 1970). We have since weight was determined by gel filtration (Andrews, demonstrated that both phenol and resorcinol are 1965) on a Sephadex G-150 column metabolized to 3-oxoadipate (Gaal & Neujahr, (2.5cm x 85cm) equilibrated with 50mM-Tris/ 1979). The metabolism of resorcinol goes through H2SO4 buffer, pH 7.6. Alcohol dehydrogenase 1,2,4-trihydroxybenzene and maleylacetate. The (mol.wt. 150000), lactate dehydrogenase (mol.wt. enzyme catalysing the reduction of maleylacetate to 136000), hexokinase (mol.wt. 105000) and avidin 3-oxoadipate was enriched from crude extracts (mol.wt. -

Xuejun Yu Dissertation Final
UNIVERSITY OF CALIFORNIA RIVERSIDE Conversion of Carbon Dioxide to Formate by a Formate Dehydrogenase from Cupriavidus necator A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Bioengineering by Xuejun Yu September 2018 Dissertation Committee: Dr. Ashok Mulchandani, Co-Chairperson Dr. Xin Ge, Co-Chairperson Dr. Russ Hille Copyright by Xuejun Yu 2018 The Dissertation of Xuejun Yu is approved: Committee Co-Chairperson Committee Co-Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to express sincere appreciation to my advisor, Professor Ashok Mulchandani, for accepting me to be his student when I was suffering. Thank you very much for your delicate guidance, support and encouragement during the past years. You not only guide me on my PhD study, but also help me to be mature on my personality. From bottom of my heart, I feel very lucky to have you as professor. Also, many thanks go to my other committee members. Professor Xin Ge acted as my co-advisor and provided many valuable comments on my molecular cloning work. Professor Russ Hille has also provided insights, encouragements and advices as a member of my committee members. I have learned so many important insights from our meetings and discussions and thank you for bringing me into the molybdenum/tungsten enzyme conference. I am also thankful to the past and current group members, colleagues and friends - Dimitri Niks, Pankaj Ramnani, Feng Tan, Rabeay Hassan, Trupti Terse, Thien-Toan Tran, Claudia Chaves, Jia-wei Tay, Hui Wang, Pham Tung, Hilda Chan, Yingning Gao and Tynan Young. -

Journal of Chromatography
aphy & S r ep og a t r a a t m i o o r n Lilla et al., J Chromatograph Separat Techniq 2012, 3:2 h T e C c f Journal of Chromatography h DOI: 10.4172/2157-7064.1000122 o n l i a q ISSN:n 2157-7064 u r e u s o J Separation Techniques Research Article OpenOpen Access Access Structural Characterization of Transglutaminase-Catalyzed Casein Cross- Linking Sergio Lilla1,2, Gianfranco Mamone2, Maria Adalgisa Nicolai1, Lina Chianese1, Gianluca Picariello2, Simonetta Caira2 and Francesco Addeo1,2* 1Dipartimento di Scienza degli Alimenti, University of Naples “Federico II”, Parco Gussone, Portici 80055, Italy 2Istituto di Scienze dell’Alimentazione (ISA) – CNR, Via Roma 64, 83100 Avellino, Italy Abstract Microbial transglutaminase is used in the food industry to improve texture by catalyzing protein cross-linking. Casein is a well-known transglutaminase substrate, but the complete role of glutamine (Q) and lysine (K) residues in its cross-linking is not fully understood. In this study, we describe the characterization of microbial Transglutaminase -modified casein using a combination of immunological and proteomic techniques. Using 5-(biotinamido)pentylamine as an acyl acceptor probe, three Q residues of β-casein and one of αs1-casein were found to participate as acyl donors. However, no Q-residues were involved in network formation with κ-casein or αs2-casein. Q and K residues in the ε-(γ-glutamyl)lysine-isopeptide bonds β-casein were identified by nanoelectrospray tandem mass spectrometry of the proteolytic digests. This work reports our progress toward a better understanding of the function and mechanism of action of microbial transglutaminase-mediated proteins.