Using MALDI-TOF MS Analysis for Rapid Discrimination of MRSA MLST
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
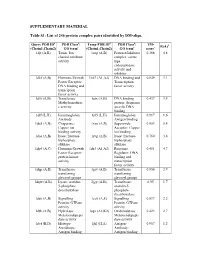
List of 246 Protein Complex Pairs Identified by DM-Align
SUPPLEMENTARY MATERIAL Table S1: List of 246 protein complex pairs identified by DM-align. Query PDB IDa PDB Classb: Temp PDB IDd PDB Classb: TM- R(Å)f (Chain1,Chain2) GO termc (Chain1,Chain2) GO termc scoree 1djt (A,B) Toxin: Ion 1aap (A,B) Protease/Inhibitor 0.368 4.8 channel inhibitor complex: serine activity type endopeptidase activity and inhibitor 1dkf (A,B) Hormone/Growth 1xb7 (A1,A2) DNA binding and 0.849 3.1 Factor Receptor: Transcription DNA binding and factor activity transciption factor activity 1dl5 (A,B) Transferase: 1utx (A,B) DNA binding 0.437 3.9 Methyltransferas protein: Sequence e activity specific DNA binding 1dlf (L,H) Immunoglobin: 1j05 (L,H) Immunoglobin: 0.917 1.6 Antibody Antigen binding 1do5 (A,B) Chaperone: 1xso (A,B) Superoxide 0.953 0.9 Copper ion Acceptor: Copper binding activity ion binding 1dos (A,B) lyase: fructose- 1rvg (A,B) lyase: fructose- 0.760 3.6 biphosphate biphosphate aldolase aldolase 1dp4 (A,C) Hormone/Growth 1dz3 (A1,A2) Response 0.481 4.7 Factor Receptor: Regulator: DNA protein kinase binding and activity transcription factor activity 1dqp (A,B) Transferase: 1grv (A,B) Transferase: 0.856 2.9 transferring transferring glycosyl groups glycosyl groups 1dqw (A,B) Lyase: orotidin- 2jgy (A,B) Transferase: 0.95 1.7 5-phosphate orotidin-5- decarboxylase phosphate decarboxylase 1ds6 (A,B) Signalling 1cc0 (A,E) Signalling 0.897 2.2 Protein: GTPase Protein: GTPase activity activity 1dth (A,B) Hydrolase: 1sqv (A2,K2) Oxidoredutase: 0.423 2.7 Metaloendopepti Metaloendopepti dase activity dase activity 1dvf -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Saccharomyces Cerevisiae Aspartate Kinase Mechanism and Inhibition
In compliance with the Canadian Privacy Legislation some supporting forms may have been removed from this dissertation. While these forms may be included in the document page count, their removal does not represent any loss of content from the dissertation. Ph.D. Thesis - D. Bareich McMaster University - Department of Biochemistry FUNGAL ASPARTATE KINASE MECHANISM AND INHIBITION By DAVID C. BAREICH, B.Sc. A Thesis Submitted to the School of Graduate Studies in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy McMaster University © Copyright by David C. Bareich, June 2003 1 Ph.D. Thesis - D. Bareich McMaster University - Department of Biochemistry FUNGAL ASPARTATE KINASE MECHANISM AND INHIBITION Ph.D. Thesis - D. Bareich McMaster University - Department of Biochemistry DOCTOR OF PHILOSOPHY (2003) McMaster University (Biochemistry) Hamilton, Ontario TITLE: Saccharomyces cerevisiae aspartate kinase mechanism and inhibition AUTHOR: David Christopher Bareich B.Sc. (University of Waterloo) SUPERVISOR: Professor Gerard D. Wright NUMBER OF PAGES: xix, 181 11 Ph.D. Thesis - D. Bareich McMaster University - Department of Biochemistry ABSTRACT Aspartate kinase (AK) from Saccharomyces cerevisiae (AKsc) catalyzes the first step in the aspartate pathway responsible for biosynthesis of L-threonine, L-isoleucine, and L-methionine in fungi. Little was known about amino acids important for AKsc substrate binding and catalysis. Hypotheses about important amino acids were tested using site directed mutagenesis to substitute these amino acids with others having different properties. Steady state kinetic parameters and pH titrations of the variant enzymes showed AKsc-K18 and H292 to be important for binding and catalysis. Little was known about how the S. cerevisiae aspartate pathway kinases, AKsc and homoserine kinase (HSKsc), catalyze the transfer of the y-phosphate from adenosine triphosphate (ATP) to L-aspartate or L-homoserine, respectively. -

Supplementary Information
Supplementary information (a) (b) Figure S1. Resistant (a) and sensitive (b) gene scores plotted against subsystems involved in cell regulation. The small circles represent the individual hits and the large circles represent the mean of each subsystem. Each individual score signifies the mean of 12 trials – three biological and four technical. The p-value was calculated as a two-tailed t-test and significance was determined using the Benjamini-Hochberg procedure; false discovery rate was selected to be 0.1. Plots constructed using Pathway Tools, Omics Dashboard. Figure S2. Connectivity map displaying the predicted functional associations between the silver-resistant gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S3. Connectivity map displaying the predicted functional associations between the silver-sensitive gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S4. Metabolic overview of the pathways in Escherichia coli. The pathways involved in silver-resistance are coloured according to respective normalized score. Each individual score represents the mean of 12 trials – three biological and four technical. Amino acid – upward pointing triangle, carbohydrate – square, proteins – diamond, purines – vertical ellipse, cofactor – downward pointing triangle, tRNA – tee, and other – circle. -

Q 297 Suppl USE
The following supplement accompanies the article Atlantic salmon raised with diets low in long-chain polyunsaturated n-3 fatty acids in freshwater have a Mycoplasma dominated gut microbiota at sea Yang Jin, Inga Leena Angell, Simen Rød Sandve, Lars Gustav Snipen, Yngvar Olsen, Knut Rudi* *Corresponding author: [email protected] Aquaculture Environment Interactions 11: 31–39 (2019) Table S1. Composition of high- and low LC-PUFA diets. Stage Fresh water Sea water Feed type High LC-PUFA Low LC-PUFA Fish oil Initial fish weight (g) 0.2 0.4 1 5 15 30 50 0.2 0.4 1 5 15 30 50 80 200 Feed size (mm) 0.6 0.9 1.3 1.7 2.2 2.8 3.5 0.6 0.9 1.3 1.7 2.2 2.8 3.5 3.5 4.9 North Atlantic fishmeal (%) 41 40 40 40 40 30 30 41 40 40 40 40 30 30 35 25 Plant meals (%) 46 45 45 42 40 49 48 46 45 45 42 40 49 48 39 46 Additives (%) 3.3 3.2 3.2 3.5 3.3 3.4 3.9 3.3 3.2 3.2 3.5 3.3 3.4 3.9 2.6 3.3 North Atlantic fish oil (%) 9.9 12 12 15 16 17 18 0 0 0 0 0 1.2 1.2 23 26 Linseed oil (%) 0 0 0 0 0 0 0 6.8 8.1 8.1 9.7 11 10 11 0 0 Palm oil (%) 0 0 0 0 0 0 0 3.2 3.8 3.8 5.4 5.9 5.8 5.9 0 0 Protein (%) 56 55 55 51 49 47 47 56 55 55 51 49 47 47 44 41 Fat (%) 16 18 18 21 22 22 22 16 18 18 21 22 22 22 28 31 EPA+DHA (% diet) 2.2 2.4 2.4 2.9 3.1 3.1 3.1 0.7 0.7 0.7 0.7 0.7 0.7 0.7 4 4.2 Table S2. -

Product Information Sheet for NR-19577
Product Information Sheet for NR-19577 Streptococcus pneumoniae Gateway® Packaging/Storage: Clone Set, Recombinant in Escherichia NR-19577 was packaged aseptically in a 96-well plate. The product is provided frozen and should be stored at -80°C or coli, Plate 10 colder immediately upon arrival. For long-term storage, the vapor phase of a liquid nitrogen freezer is recommended. Catalog No. NR-19577 Freeze-thaw cycles should be avoided. This reagent is the tangible property of the U.S. Government. Growth Conditions: For research use only. Not for human use. Media: LB broth containing 50 µg/mL kanamycin LB agar containing 50 µg/mL kanamycin Contributor: Incubation: Pathogen Functional Genomics Resource Center at the J. Temperature: 37°C Craig Venter Institute Atmosphere: Aerobic Propagation: Manufacturer: 1. Scrape top of frozen well with a pipette tip and streak BEI Resources onto agar plate. 2. Incubate the plates at 37°C for 18 to 24 hours. Product Description: Clone plates are replicated using a BioMek® FX robot. Citation: Production in the 96-well format has increased risk of cross- Acknowledgment for publications should read “The following contamination between adjacent wells. Individual clones reagent was obtained through BEI Resources, NIAID, NIH: should be purified (e.g. single colony isolation and Streptococcus pneumoniae Gateway® Clone Set, purification using good microbiological practices) and Recombinant in Escherichia coli, Plate 10, NR-19577.” sequence-verified prior to use. BEI Resources only confirms the clone plate orientation and viability of randomly picked Biosafety Level: 1 clones. BEI Resources does not confirm or validate Appropriate safety procedures should always be used with individual clone identities provided by the contributor. -

Consolidated List of Up-Regulated Proteins Expressed at Different Cr (VI) Concentrations at Time Points
Electronic Supplementary Material (ESI) for Metallomics. -

Grasnotice 946, Lactobacillus Plantarum Strain DSM 33452
GRAS Notice (GRN) No. 946 https://www.fda.gov/food/generally-recognized-safe-gras/ RECEIVED gras-notice-inventory JUNO 2 1020 Chr. Hansen, In -?FFICE OF FOOD ADD ITIVE SAFETY 9015 West Maple Street Milwaukee, WI 53214 - 4298 Division of Biotechnology and GRAS Notice Review U.S .A. Center for Food Safety & Applied Nutrition (HFS-255) U.S. Food & Drug Administration Phone 414 - 607 - 5700 Fax 414 - 607 - 5959 Reference: Lactobacillus plantarum DSM 33452 June 1, 2020 Dear Sir or Madam, In accordance with the Federal Register [81 Fed. Reg. 159 (17 August 2016)] issuance on Generally Recognized as Safe (GRAS) notifications (21 CFR Part 170), Chr. Hansen is pleased to submit a notice that we have concluded, through scientific procedures that Lactobacillus plantarum (L. plantarum) DSM 33452 is generally recognized as safe for use in malolactic fermentation of wine and musts and is not subject to the pre-market approval requirements. The recommendation is to inoculate the pure starter culture of L. plantarum DSM 33452 into wine or must at an inoculation level of 1.0E+07 CFU/g at the time of crushing grapes or as early as possible in the fermentation tanks. L. plantarum DSM 33452 is sensitive to alcohol concentration, so the concentration of the organism will decrease as the concentration of alcohol in the wine or must increases. Though L. plantarum DSM 33452 is safe to consume, it would be present at negligible levels, if at all, in the finished product. It should also be noted that due to recent taxonomic changes to the genus Lactobaci/lus, Lactobacillus plantarum will be known as Lactiplantibacillus plantarum moving forward (Zheng, et al., 2020). -

Assay for Detecting Mycoplasma by Measuring Acetate Kinase Or Carbamate Kinase Activity
(19) & (11) EP 2 264 181 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 22.12.2010 Bulletin 2010/51 C12Q 1/04 (2006.01) C12Q 1/48 (2006.01) C12Q 1/00 (2006.01) (21) Application number: 10075423.3 (22) Date of filing: 16.04.2004 (84) Designated Contracting States: • Crouch, Sharon Patricia Mary AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Nottingham NG7 1DQ (GB) HU IE IT LI LU MC NL PL PT RO SE SI SK TR • Slater, Kevin John Nottingham NG7 1ED (GB) (30) Priority: 17.04.2003 GB 0308829 • Cox, Anne 17.04.2003 US 436323 P Long Eaton, Nottingham NG10 3JB (GB) (62) Document number(s) of the earlier application(s) in (74) Representative: Didmon, Mark et al accordance with Art. 76 EPC: Potter Clarkson LLP 04727944.3 / 1 613 763 Park View House 58 The Ropewalk (71) Applicant: Cambrex Bio Science Nottingham Nottingham NG1 5DD (GB) Limited Nottingham NG1 1GF (GB) Remarks: This application was filed on 13-09-2010 as a (72) Inventors: divisional application to the application mentioned • Pitt, Anthony under INID code 62. Beeston, Nottingham NG9 2BE (GB) (54) ASSAY FOR DETECTING MYCOPLASMA BY MEASURING ACETATE KINASE OR CARBAMATE KINASE ACTIVITY (57) The invention relates to a method of detecting (ii) detecting and/or measuring the activity of acetate ki- the presence of mycoplasma in a test sample comprising: nase and/or carbamate kinase in the test sample, the (i) providing a test sample; and activity being indicative of contamination by mycoplas- ma. -

The Carbamate Kinase-Like Carbamoyl Phosphate Synthetase of The
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 12803–12808, November 1997 Biochemistry The carbamate kinase-like carbamoyl phosphate synthetase of the hyperthermophilic archaeon Pyrococcus furiosus, a missing link in the evolution of carbamoyl phosphate biosynthesis VIRGINIE DURBECQ*, CHRISTIANNE LEGRAIN†,MARTINE ROOVERS‡§,ANDRE´ PIE´RARD*†, AND NICOLAS GLANSDORFF†‡§¶ *Laboratoire de Microbiologie, Universite´Libre de Bruxelles, †Research Institute of the Centre de Recherches et d’Enseignement des Industries Alimentaires et Chimiques, ‡Microbiologie, Vrije Universiteit Brussel, and §Flanders Interuniversity Institute for Biotechnology, Avenue E. Gryson 1, B-1070 Brussels, Belgium Communicated by H. Edwin Umbarger, Purdue University, West Lafayette, IN, September 16, 1997 (received for review July 9, 1997) ABSTRACT Microbial carbamoyl phosphate synthetases protein, and it was proposed that the synthetase gene had (CPS) use glutamine as nitrogen donor and are composed of evolved by duplication of a smaller ancestral gene—possibly two subunits (or domains), one exhibiting glutaminase activ- coding for a carbamate kinase (CK) (5). CKs (EC 2.7.2.2) are ity, the other able to synthesize carbamoyl phosphate (CP) homodimers composed of subunits of 31–37 kDa (6). As from bicarbonate, ATP, and ammonia. The pseudodimeric carbamate is in chemical equilibrium with bicarbonate and 2 organization of this synthetase suggested that it has evolved by ammonia, CK can synthesize CP from HCO3 ,NH3 and one duplication of a smaller kinase, possibly a carbamate kinase molecule of ATP (Fig. 1). This evolutionary scheme found (CK). In contrast to other prokaryotes the hyperthermophilic support in the weak but nevertheless suggestive sequence archaeon Pyrococcus furiosus was found to synthesize CP by similarities that were brought to light between carB and the using ammonia and not glutamine. -
Sigma Enzymes, Coenzymes and Enzyme Substrates
Sigma Enzymes, Coenzymes and Enzyme Substrates Library Listing – 485 spectra This library represents a material-specific subset of the larger Sigma Biochemical Condensed Phase Library relating to enzymes, coenzymes and enzyme substrates found in the Sigma Biochemicals and Reagents catalog. Spectra acquired by Sigma-Aldrich Co. which were examined and processed at Thermo Fisher Scientific. The spectra include compound name, molecular formula, CAS (Chemical Abstract Service) registry number, and Sigma catalog number. Sigma Enzymes, Coenzymes and Enzyme Substrates Index Compound Name Index Compound Name 484 (E)-Vaccenoyl coenzyme A 298 Azo soybean flour 483 (Z)-Vaccenoyl coenzyme A 295 Azoalbumin 400 1,N6-Etheno acetyl coenzyme A, Li salt 296 Azocasein 401 1,N6-Etheno coenzyme A, Na + Li salt 297 Azocoll 397 11,14,17-Eicosatrienoyl coenzyme A 379 Behenoyl coenzyme A 396 11,14-Eicosadienoyl coenzyme A 380 Benzoyl coenzyme A, Li salt 395 11-Eicosaenoyl coenzyme A 343 Bis-(p-nitrophenyl) phosphate 321 2-(b-D-Galactosidoxy)naphthol AS-LC 344 Bis-(p-nitrophenyl) phosphate, Ca salt 394 3'-Dephosphocoenzyme A 345 Bis-(p-nitrophenyl) phosphate, Na salt 370 3-Acetylpyridine adenine dinucleotide 381 Brassidoyl coenzyme A 372 3-Acetylpyridine adenine dinucleotide 9 Carbamate kinase from streptococcus phosphate, Na salt faecalis 371 3-Acetylpyridine adenine dinucleotide, 310 Carbamyl phosphate, diammonium salt reduced form 311 Carbamyl phosphate, dilithium salt 373 3-Acetylpyridine-hypoxanthine 312 Carbamyl phosphate, disodium salt dinucleotide 10 Carbonic -
BMC Structural Biology Biomed Central
BMC Structural Biology BioMed Central Research article Open Access A comprehensive update of the sequence and structure classification of kinases Sara Cheek2, Krzysztof Ginalski2,3, Hong Zhang2 and Nick V Grishin*1,2 Address: 1Howard Hughes Medical Institute, University of Texas Southwestern Medical Center 5323 Harry Hines Blvd., Dallas, Texas 75390, USA, 2Department of Biochemistry, University of Texas Southwestern Medical Center 5323 Harry Hines Blvd., Dallas, Texas 75390, USA and 3Bioinformatics Laboratory, Interdisciplinary Centre for Mathematical and Computational Modelling Warsaw University, Pawinskiego 5a, 02-106 Warsaw, Poland Email: Sara Cheek - [email protected]; Krzysztof Ginalski - [email protected]; Hong Zhang - [email protected]; Nick V Grishin* - [email protected] * Corresponding author Published: 16 March 2005 Received: 11 January 2005 Accepted: 16 March 2005 BMC Structural Biology 2005, 5:6 doi:10.1186/1472-6807-5-6 This article is available from: http://www.biomedcentral.com/1472-6807/5/6 © 2005 Cheek et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: A comprehensive update of the classification of all available kinases was carried out. This survey presents a complete global picture of this large functional class of proteins and confirms the soundness of our initial kinase classification scheme. Results: The new survey found the total number of kinase sequences in the protein database has increased more than three-fold (from 17,310 to 59,402), and the number of determined kinase structures increased two-fold (from 359 to 702) in the past three years.