Supplemental Table A. Predicted Function and Classification of All Mmymysc Orfs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Electronic Supplementary Material (ESI) for Analyst. This Journal Is © the Royal Society of Chemistry 2017
Electronic Supplementary Material (ESI) for Analyst. This journal is © The Royal Society of Chemistry 2017 Supplemental Table 2. Proteins Increased in Either Blood or Horizon media Table 2A. Proteins Increased in Spores Produced on Horizon Soil Over Spores Produced on Blood Medium quasi.fdr Protein Protein Class/Name KEGG Pathway Names or Function (if ID Pathways found in KEGG) Amino Acid Metabolism bat00250, bat00280, Alanine, aspartate and glutamate metabolism, bat00410, Valine, leucine and isoleucine degradation, bat00640, beta-Alanine metabolism to acetyl CoA, 4.50E-07 BAS0310 4-aminobutyrate aminotransferase bat00650 Propanoate metabolism, Butanoate metabolism bat00270, bat00330, Cysteine and methionine metabolism, Arginine bat00410, and proline metabolism, beta-Alanine 3.84E-10 BAS5060 spermidine synthase bat00480 metabolism, Glutathione metabolism bat00270, bat00330, Cysteine and methionine metabolism, Arginine bat00410, and proline metabolism, beta-Alanine 2.30E-09 BAS5219 spermidine synthase bat00480 metabolism, Glutathione metabolism bat00250, Alanine, aspartate and glutamate metabolism, 6.94E-07 BAS0561 alanine dehydrogenase bat00430 Taurine and hypotaurine metabolism bat00250, Alanine, aspartate and glutamate metabolism, 5.23E-07 BAS4521 alanine dehydrogenase bat00430 Taurine and hypotaurine metabolism 0.005627 BAS5218 agmatinase, putative bat00330 Arginine and proline metabolism 2,3,4,5-tetrahydropyridine-2- carboxylate N-succinyltransferase, 1.40E-10 BAS3891 putative bat00300 Lysine biosynthesis bat00010, bat00020, Glycolysis -

Articles Catalytic Cycling in Β-Phosphoglucomutase: a Kinetic
9404 Biochemistry 2005, 44, 9404-9416 Articles Catalytic Cycling in â-Phosphoglucomutase: A Kinetic and Structural Analysis†,‡ Guofeng Zhang, Jianying Dai, Liangbing Wang, and Debra Dunaway-Mariano* Department of Chemistry, UniVersity of New Mexico, Albuquerque, New Mexico 87131-0001 Lee W. Tremblay and Karen N. Allen* Department of Physiology and Biophysics, Boston UniVersity School of Medicine, Boston, Massachusetts 02118-2394 ReceiVed March 26, 2005; ReVised Manuscript ReceiVed May 18, 2005 ABSTRACT: Lactococcus lactis â-phosphoglucomutase (â-PGM) catalyzes the interconversion of â-D-glucose 1-phosphate (â-G1P) and â-D-glucose 6-phosphate (G6P), forming â-D-glucose 1,6-(bis)phosphate (â- G16P) as an intermediate. â-PGM conserves the core domain catalytic scaffold of the phosphatase branch of the HAD (haloalkanoic acid dehalogenase) enzyme superfamily, yet it has evolved to function as a mutase rather than as a phosphatase. This work was carried out to identify the structural basis underlying this diversification of function. In this paper, we examine â-PGM activation by the Mg2+ cofactor, â-PGM activation by Asp8 phosphorylation, and the role of cap domain closure in substrate discrimination. First, the 1.90 Å resolution X-ray crystal structure of the Mg2+-â-PGM complex is examined in the context of + + previously reported structures of the Mg2 -R-D-galactose-1-phosphate-â-PGM, Mg2 -phospho-â-PGM, and Mg2+-â-glucose-6-phosphate-1-phosphorane-â-PGM complexes to identify conformational changes that occur during catalytic turnover. The essential role of Asp8 in nucleophilic catalysis was confirmed by demonstrating that the D8A and D8E mutants are devoid of catalytic activity. -

Mycobacterium Haemophilum Sp. Nov., a New Pathogen of Humanst
0020-7713/78/0028-0067$02.0/0 INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, Jan. 1978, p. 67-75 Vol. 28, No. 1 Copyright 0 1978 International Association of Microbiological Societies Printed in U.S. A. Mycobacterium haemophilum sp. nov., a New Pathogen of Humanst DAVID SOMPOLINSKY,’v2 ANNIE LAGZIEL,’ DAVID NAVEH,3 AND TULI YANKILEVITZ3 Department of Microbwlogy, Asaf Harofe Government Hospital, Zerifin‘; Rapaport Laboratories, Bar-Ilan University, Ramat-Gan2;and Department of Internal Medicine “A,”Meir Hospital, Kfar Saba,3 Israel A patient under immunosuppressive treatment of Hodgkin’s disease developed generalized skin granulomata and subcutaneous abscesses. Several aspirated pus samples yielded acid-fast rods with the following properties: temperature opti- mum, about 30°C with no growth at 37°C; slow growth (2 to 4 weeks); nonchrom- ogenic; hemoglobin or hemin requirement for growth; catalase negative; pyrazin- amidase and nicotinamidase positive; and urease negative. The guanine-plus- cytosine content of the deoxyribonucleic acid was calculated from the melting temperature to be 66.0 mol%. It is concluded that these isolates belong to a new species, for which the name Mycobacterium haemophilum is proposed. The type strain of this species is strain 1 (= ATCC 29548). The new species is related to M. marinum and M. ulcerans. Granulomatous skin diseases of humans CASE HISTORY caused by mycobacteria other than Mycobacte- After World War 11, a 27-year-old woman was di- rium tuberculosis and M. leprae are well known. agnosed as having tuberculosis. She received treat- The two organisms most often involved etiolog- ment until 1951. In February 1969, at the age of 51, ically are M. -

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Pseudouridine Synthase 1: a Site-Specific Synthase Without Strict Sequence Recognition Requirements Bryan S
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Published online 18 November 2011 Nucleic Acids Research, 2012, Vol. 40, No. 5 2107–2118 doi:10.1093/nar/gkr1017 Pseudouridine synthase 1: a site-specific synthase without strict sequence recognition requirements Bryan S. Sibert and Jeffrey R. Patton* Department of Pathology, Microbiology and Immunology, University of South Carolina, School of Medicine, Columbia, SC 29208 USA Received May 20, 2011; Revised October 19, 2011; Accepted October 22, 2011 ABSTRACT rRNA and snRNA and requires Dyskerin or its homologs Pseudouridine synthase 1 (Pus1p) is an unusual (Cbf5p in yeast for example) and RNP cofactors [most site-specific modification enzyme in that it can often H/ACA small nucleolar ribonucleoprotein particles modify a number of positions in tRNAs and can rec- (snoRNPs)] that enable one enzyme to recognize many ognize several other types of RNA. No consensus different sites for modification on different substrates recognition sequence or structure has been identi- (17–25). The other pathway for É formation employs fied for Pus1p. Human Pus1p was used to determine site-specific É synthases that require no cofactors to rec- which structural or sequence elements of human ognize and modify the RNA substrate. A number of en- tRNASer are necessary for pseudouridine ()) forma- zymes have been identified in this pathway and are grouped in six families that all share a common basic tion at position 28 in the anticodon stem-loop (ASL). Ser structure (4). It is safe to say the cofactor ‘guided’ pathway Some point mutations in the ASL stem of tRNA has received a great deal of attention because of its simi- had significant effects on the levels of modification larity to aspects of RNA editing, but the site-specific and compensatory mutation, to reform the base pseudouridine synthases accomplish the same task, on pair, restored a wild-type level of ) formation. -

Polymerase Ribozyme with Promoter Recognition
In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition by Razvan Cojocaru BSc, Simon Fraser University, 2014 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Department of Molecular Biology and Biochemistry Faculty of Science © Razvan Cojocaru 2021 SIMON FRASER UNIVERSITY Summer 2021 Copyright in this work is held by the author. Please ensure that any reproduction or re-use is done in accordance with the relevant national copyright legislation. Declaration of Committee Name: Razvan Cojocaru Degree: Doctor of Philosophy Title: In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition Committee: Chair: Lisa Craig Professor, Molecular Biology and Biochemistry Peter Unrau Supervisor Professor, Molecular Biology and Biochemistry Dipankar Sen Committee Member Professor, Molecular Biology and Biochemistry Michel Leroux Committee Member Professor, Molecular Biology and Biochemistry Mani Larijani Internal Examiner Associate Professor, Molecular Biology and Biochemistry Gerald Joyce External Examiner Professor, Jack H. Skirball Center for Chemical Biology and Proteomics Salk Institute for Biological Studies Date Defended/Approved: August 12, 2021 ii Abstract The RNA World hypothesis proposes that the early evolution of life began with RNAs that can serve both as carriers of genetic information and as catalysts. Later in evolution, these functions were gradually replaced by DNA and enzymatic proteins in cellular biology. I start by reviewing the naturally occurring catalytic RNAs, ribozymes, as they play many important roles in biology today. These ribozymes are central to protein synthesis and the regulation of gene expression, creating a landscape that strongly supports an early RNA World. -

Diagnosis, Treatment and Follow Up
DOI: 10.1002/jimd.12024 REVIEW International clinical guidelines for the management of phosphomannomutase 2-congenital disorders of glycosylation: Diagnosis, treatment and follow up Ruqaiah Altassan1,2 | Romain Péanne3,4 | Jaak Jaeken3 | Rita Barone5 | Muad Bidet6 | Delphine Borgel7 | Sandra Brasil8,9 | David Cassiman10 | Anna Cechova11 | David Coman12,13 | Javier Corral14 | Joana Correia15 | María Eugenia de la Morena-Barrio16 | Pascale de Lonlay17 | Vanessa Dos Reis8 | Carlos R Ferreira18,19 | Agata Fiumara5 | Rita Francisco8,9,20 | Hudson Freeze21 | Simone Funke22 | Thatjana Gardeitchik23 | Matthijs Gert4,24 | Muriel Girad25,26 | Marisa Giros27 | Stephanie Grünewald28 | Trinidad Hernández-Caselles29 | Tomas Honzik11 | Marlen Hutter30 | Donna Krasnewich18 | Christina Lam31,32 | Joy Lee33 | Dirk Lefeber23 | Dorinda Marques-da-Silva9,20 | Antonio F Martinez34 | Hossein Moravej35 | Katrin Õunap36,37 | Carlota Pascoal8,9 | Tiffany Pascreau38 | Marc Patterson39,40,41 | Dulce Quelhas14,42 | Kimiyo Raymond43 | Peymaneh Sarkhail44 | Manuel Schiff45 | Małgorzata Seroczynska29 | Mercedes Serrano46 | Nathalie Seta47 | Jolanta Sykut-Cegielska48 | Christian Thiel30 | Federic Tort27 | Mari-Anne Vals49 | Paula Videira20 | Peter Witters50,51 | Renate Zeevaert52 | Eva Morava53,54 1Department of Medical Genetic, Montréal Children's Hospital, Montréal, Québec, Canada 2Department of Medical Genetic, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia 3Department of Human Genetics, KU Leuven, Leuven, Belgium 4LIA GLYCOLAB4CDG (International -

| Hai Lui a Un Acutul Luniit Moonhiti
|HAI LUI AUN ACUTULUS010006055B2 LUNIIT MOONHITI (12 ) United States Patent (10 ) Patent No. : US 10 , 006 , 055 B2 Burk et al. (45 ) Date of Patent: Jun . 26 , 2018 ( 54 ) MICROORGANISMS FOR PRODUCING 2002/ 0168654 A1 11/ 2002 Maranas et al. 2003 / 0059792 Al 3 /2003 Palsson et al . BUTADIENE AND METHODS RELATED 2003 /0087381 A1 5 / 2003 Gokarn THERETO 2003 / 0224363 Al 12 /2003 Park et al . 2003 / 0233218 Al 12 /2003 Schilling (71 ) Applicant: Genomatica , Inc. , San Diego , CA (US ) 2004 / 0009466 AL 1 /2004 Maranas et al. 2004 / 0029149 Al 2 /2004 Palsson et al. ( 72 ) Inventors : Mark J . Burk , San Diego , CA (US ) ; 2004 / 0072723 A1 4 /2004 Palsson et al. Anthony P . Burgard , Bellefonte , PA 2004 / 0152159 Al 8 / 2004 Causey et al . 2005 /0042736 A1 2 / 2005 San et al . (US ) ; Robin E . Osterhout , San Diego , 2005 / 0079482 A1 4 / 2005 Maranas et al . CA (US ) ; Jun Sun , San Diego , CA 2006 / 0046288 Al 3 / 2006 Ka - Yiu et al. ( US ) ; Priti Pharkya , San Diego , CA 2006 / 0073577 A1 4 / 2006 Ka - Yiu et al . (US ) 2007 /0184539 Al 8 / 2007 San et al . 2009 / 0047718 Al 2 / 2009 Blaschek et al . 2009 / 0047719 Al 2 / 2009 Burgard et al . (73 ) Assignee : Genomatica , Inc ., San Diego , CA (US ) 2009 /0191593 A1 7 / 2009 Burk et al . 2010 / 0003716 A1 1 / 2010 Cervin et al. ( * ) Notice : Subject to any disclaimer , the term of this 2010 /0184171 Al 7 /2010 Jantama et al. patent is extended or adjusted under 35 2010 /0304453 Al 12 / 2010 Trawick et al . -

Genome of Phaeocystis Globosa Virus Pgv-16T Highlights the Common Ancestry of the Largest Known DNA Viruses Infecting Eukaryotes
Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes Sebastien Santinia, Sandra Jeudya, Julia Bartolia, Olivier Poirota, Magali Lescota, Chantal Abergela, Valérie Barbeb, K. Eric Wommackc, Anna A. M. Noordeloosd, Corina P. D. Brussaardd,e,1, and Jean-Michel Claveriea,f,1 aStructural and Genomic Information Laboratory, Unité Mixte de Recherche 7256, Centre National de la Recherche Scientifique, Aix-Marseille Université, 13288 Marseille Cedex 9, France; bCommissariat à l’Energie Atomique–Institut de Génomique, 91057 Evry Cedex, France; cDepartment of Plant and Soil Sciences, University of Delaware, Newark, DE 19711; dDepartment of Biological Oceanography, Royal Netherlands Institute for Sea Research, NL-1790 AB Den Burg (Texel), The Netherlands; eAquatic Microbiology, Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, The Netherlands; and fService de Santé Publique et d’Information Médicale, Hôpital de la Timone, Assistance Publique–Hôpitaux de Marseille, FR-13385 Marseille, France Edited by James L. Van Etten, University of Nebraska, Lincoln, NE, and approved May 1, 2013 (received for review February 22, 2013) Large dsDNA viruses are involved in the population control of many viruses: 730 kb and 1.28 Mb for CroV and Megavirus chilensis, globally distributed species of eukaryotic phytoplankton and have respectively. Other studies, targeting virus-specific genes [e.g., a prominent role in bloom termination. The genus Phaeocystis (Hap- DNA polymerase B (8) or capsid proteins (9)] have suggested tophyta, Prymnesiophyceae) includes several high-biomass-forming a close phylogenetic relationship between Mimivirus and several phytoplankton species, such as Phaeocystis globosa, the blooms of giant dsDNA viruses infecting various unicellular algae such as which occur mostly in the coastal zone of the North Atlantic and the Pyramimonas orientalis (Chlorophyta, Prasinophyceae), Phaeocys- North Sea. -

Phosphatidylinositol-3-Kinase Related Kinases (Pikks) in Radiation-Induced Dna Damage
Mil. Med. Sci. Lett. (Voj. Zdrav. Listy) 2012, vol. 81(4), p. 177-187 ISSN 0372-7025 DOI: 10.31482/mmsl.2012.025 REVIEW ARTICLE PHOSPHATIDYLINOSITOL-3-KINASE RELATED KINASES (PIKKS) IN RADIATION-INDUCED DNA DAMAGE Ales Tichy 1, Kamila Durisova 1, Eva Novotna 1, Lenka Zarybnicka 1, Jirina Vavrova 1, Jaroslav Pejchal 2, Zuzana Sinkorova 1 1 Department of Radiobiology, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic 2 Centrum of Advanced Studies, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic. Received 5 th September 2012. Revised 27 th November 2012. Published 7 th December 2012. Summary This review describes a drug target for cancer therapy, family of phosphatidylinositol-3 kinase related kinases (PIKKs), and it gives a comprehensive review of recent information. Besides general information about phosphatidylinositol-3 kinase superfamily, it characterizes a DNA-damage response pathway since it is monitored by PIKKs. Key words: PIKKs; ATM; ATR; DNA-PK; Ionising radiation; DNA-repair ABBREVIATIONS therapy and radiation play a pivotal role. Since cancer is one of the leading causes of death worldwide, it is DSB - double stand breaks, reasonable to invest time and resources in the enligh - IR - ionising radiation, tening of mechanisms, which underlie radio-resis - p53 - TP53 tumour suppressors, tance. PI - phosphatidylinositol. The aim of this review is to describe the family INTRODUCTION of phosphatidyinositol 3-kinases (PI3K) and its func - tional subgroup - phosphatidylinositol-3-kinase rela - An efficient cancer treatment means to restore ted kinases (PIKKs) and their relation to repairing of controlled tissue growth via interfering with cell sig - radiation-induced DNA damage. -
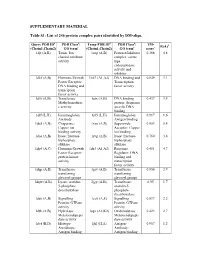
List of 246 Protein Complex Pairs Identified by DM-Align
SUPPLEMENTARY MATERIAL Table S1: List of 246 protein complex pairs identified by DM-align. Query PDB IDa PDB Classb: Temp PDB IDd PDB Classb: TM- R(Å)f (Chain1,Chain2) GO termc (Chain1,Chain2) GO termc scoree 1djt (A,B) Toxin: Ion 1aap (A,B) Protease/Inhibitor 0.368 4.8 channel inhibitor complex: serine activity type endopeptidase activity and inhibitor 1dkf (A,B) Hormone/Growth 1xb7 (A1,A2) DNA binding and 0.849 3.1 Factor Receptor: Transcription DNA binding and factor activity transciption factor activity 1dl5 (A,B) Transferase: 1utx (A,B) DNA binding 0.437 3.9 Methyltransferas protein: Sequence e activity specific DNA binding 1dlf (L,H) Immunoglobin: 1j05 (L,H) Immunoglobin: 0.917 1.6 Antibody Antigen binding 1do5 (A,B) Chaperone: 1xso (A,B) Superoxide 0.953 0.9 Copper ion Acceptor: Copper binding activity ion binding 1dos (A,B) lyase: fructose- 1rvg (A,B) lyase: fructose- 0.760 3.6 biphosphate biphosphate aldolase aldolase 1dp4 (A,C) Hormone/Growth 1dz3 (A1,A2) Response 0.481 4.7 Factor Receptor: Regulator: DNA protein kinase binding and activity transcription factor activity 1dqp (A,B) Transferase: 1grv (A,B) Transferase: 0.856 2.9 transferring transferring glycosyl groups glycosyl groups 1dqw (A,B) Lyase: orotidin- 2jgy (A,B) Transferase: 0.95 1.7 5-phosphate orotidin-5- decarboxylase phosphate decarboxylase 1ds6 (A,B) Signalling 1cc0 (A,E) Signalling 0.897 2.2 Protein: GTPase Protein: GTPase activity activity 1dth (A,B) Hydrolase: 1sqv (A2,K2) Oxidoredutase: 0.423 2.7 Metaloendopepti Metaloendopepti dase activity dase activity 1dvf