Sigma Enzymes, Coenzymes and Enzyme Substrates
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Proteomic Analysis of Mycelial Proteins from Magnaporthe Oryzae Under Nitrogen Starvation
Proteomic analysis of mycelial proteins from Magnaporthe oryzae under nitrogen starvation X.-G. Zhou1,2, P. Yu 2, C. Dong2, C.-X. Yao2, Y.-M. Ding2, N. Tao2 and Z.-W. Zhao1 1State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, Kunming, China 2Key Laboratory of Southwestern Crop Gene Resources and Germplasm Innovation, Ministry of Agriculture and Yunnan Provincial Key Laboratory of Agricultural Biotechnology, Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences, Kunming, China Corresponding author: Z.W. Zhao E-mail: [email protected] Genet. Mol. Res. 15 (2): gmr.15028637 Received March 23, 2016 Accepted April 11, 2016 Published May 13, 2016 DOI http://dx.doi.org/10.4238/gmr.15028637 ABSTRACT. Magnaporthe oryzae is an important model system in studies of plant pathogenic fungi, and nitrogen is a key nutrient source affecting microbial growth and development. In order to understand how nitrogen stress causes changes in mycelial proteins, we analyzed differentially expressed mycelial proteins from the M. oryzae virulent strain CH-63 using two-dimensional electrophoresis and mass spectrometry in complete medium or under nitrogen starvation conditions. A total of 975 ± 70 and 1169 ± 90 protein spots were detected in complete medium and under nitrogen starvation conditions, respectively. Forty-nine protein spots exhibited at least 2-fold up- regulation or down-regulation at the protein level according to PDQuest7.4. Moreover, 43 protein spots were successfully identified by matrix-assisted laser desorption/ionization-time-of-flight/time-of-flight mass spectrometry. Among these spots, 6 proteins were functionally unknown and 37 proteins were categorized into 5 groups according to Genetics and Molecular Research 15 (2): gmr.15028637 ©FUNPEC-RP www.funpecrp.com.br X.-G. -

Updating the Sequence-Based Classification of Glycosyl Hydrolases
Article Updating the sequence-based classification of glycosyl hydrolases HENRISSAT, Bernard, BAIROCH, Amos Marc Reference HENRISSAT, Bernard, BAIROCH, Amos Marc. Updating the sequence-based classification of glycosyl hydrolases. Biochemical Journal, 1996, vol. 316 ( Pt 2), p. 695-6 PMID : 8687420 DOI : 10.1042/bj3160695 Available at: http://archive-ouverte.unige.ch/unige:36909 Disclaimer: layout of this document may differ from the published version. 1 / 1 Biochem. J. (1996) 316, 695–696 (Printed in Great Britain) 695 BIOCHEMICAL JOURNAL Updating the sequence-based classification of available. When the number of glycosyl hydrolase sequences reached C 480, ten additional families (designated 36–45) could glycosyl hydrolases be defined and were added to the classification [2]. There are at present over 950 sequences of glycosyl hydrolases in the data- A classification of glycosyl hydrolases based on amino-acid- banks (EMBL}GenBank and SWISS-PROT). Their analysis sequence similarities was proposed in this Journal a few years shows that the vast majority of the C 470 additional sequences ago [1]. This classification originated from the analysis of C 300 that have become available since the last update could be classified sequences and their grouping into 35 families designated 1–35. in the existing families. However, several sequences not fitting Because such a classification is necessarily sensitive to the sample, the existing families allow the definition of new families (desig- it was anticipated that it was incomplete and that new families nated 46–57) (Table 1). When the several present genome would be determined when additional sequences would become sequencing projects have reached completion, the number of Table 1 New families in the classification of glycosyl hydrolases Family Enzyme Organism SWISS-PROT EMBL/GenBank 46 Chitosanase Bacillus circulans MH-K1 P33673 D10624 46 Chitosanase Streptomyces sp. -

Oxdc Antibody Rabbit Polyclonal Antibody Catalog # ABV11223
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 OxdC Antibody Rabbit Polyclonal Antibody Catalog # ABV11223 Specification OxdC Antibody - Product Information Application WB Primary Accession O34714 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit IgG Calculated MW 43566 OxdC Antibody - Additional Information Gene ID 938620 Positive Control Western Blot: Recombinant protein Application & Usage Western blot: 1-4 Western blot of Oxalate decarboxylase µg/ml. antibody. Lane 1: rb- Oxalate decarboxylase - Other Names 10 ng. Lane 2: rb- Oxalate decarboxylase - 50 YvrK ng Target/Specificity OxdC OxdC Antibody - Background Antibody Form Oxalate decarboxylase (OxdC, EC4.1.1.2) is a Liquid manganese-containing enzyme, which decomposes oxalic acid and oxalate. With Appearance OxdC catalysis, oxalate is split into formate Colorless liquid and CO2. This enzyme belongs to the family of lyases, specifically the carboxy-lyases, which Formulation 100 µg (0.5 mg/ml) of antibody in PBS pH cleave carbon-carbon bonds. The systematic 7.2 containing 0.01 % BSA, 0.01 % name of this enzyme class is oxalate thimerosal, and 50 % glycerol. carboxy-lyase (formate-forming). This enzyme is also called oxalate carboxy-lyase. The Handling enzyme is composed of two cupin domains, The antibody solution should be gently each of which contains a Mn (II) ion. This mixed before use. enzyme participates in glyoxylate and dicarboxylate metabolism. This enzyme has Reconstitution & Storage been recognized for diagnostics in diverse -20 °C biotechnological applications such as the clinical assay of oxalate in blood and urine, Background Descriptions therapeutics, process industry, and agriculture to lower oxalate levels in foods and the environment. -

Proteo-Metabolomic Investigation of Transgenic Rice Unravels Metabolic
www.nature.com/scientificreports OPEN Proteo-metabolomic investigation of transgenic rice unravels metabolic alterations and Received: 27 November 2018 Accepted: 24 June 2019 accumulation of novel proteins Published: xx xx xxxx potentially involved in defence against Rhizoctonia solani Subhasis Karmakar1, Karabi Datta1, Kutubuddin Ali Molla2,3, Dipak Gayen4, Kaushik Das1, Sailendra Nath Sarkar1 & Swapan K. Datta1 The generation of sheath blight (ShB)-resistant transgenic rice plants through the expression of Arabidopsis NPR1 gene is a signifcant development for research in the feld of biotic stress. However, to our knowledge, regulation of the proteomic and metabolic networks in the ShB-resistant transgenic rice plants has not been studied. In the present investigation, the relative proteome and metabolome profles of the non–transformed wild-type and the AtNPR1-transgenic rice lines prior to and subsequent to the R. solani infection were investigated. Total proteins from wild type and transgenic plants were investigated using two-dimensional gel electrophoresis (2-DE) followed by mass spectrometry (MS). The metabolomics study indicated an increased abundance of various metabolites, which draws parallels with the proteomic analysis. Furthermore, the proteome data was cross-examined using network analysis which identifed modules that were rich in known as well as novel immunity-related prognostic proteins, particularly the mitogen-activated protein kinase 6, probable protein phosphatase 2C1, probable trehalose-phosphate phosphatase 2 and heat shock protein. A novel protein, 14–3– 3GF14f was observed to be upregulated in the leaves of the transgenic rice plants after ShB infection, and the possible mechanistic role of this protein in ShB resistance may be investigated further. -

To Study Mutant P53 Gain of Function, Various Tumor-Derived P53 Mutants
Differential effects of mutant TAp63γ on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression. A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By Shama K Khokhar M.Sc., Bilaspur University, 2004 B.Sc., Bhopal University, 2002 2007 1 COPYRIGHT SHAMA K KHOKHAR 2007 2 WRIGHT STATE UNIVERSITY SCHOOL OF GRADUATE STUDIES Date of Defense: 12-03-07 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY SHAMA KHAN KHOKHAR ENTITLED Differential effects of mutant TAp63γ on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science Madhavi P. Kadakia, Ph.D. Thesis Director Daniel Organisciak , Ph.D. Department Chair Committee on Final Examination Madhavi P. Kadakia, Ph.D. Steven J. Berberich, Ph.D. Michael Leffak, Ph.D. Joseph F. Thomas, Jr., Ph.D. Dean, School of Graduate Studies 3 Abstract Khokhar, Shama K. M.S., Department of Biochemistry and Molecular Biology, Wright State University, 2007 Differential effect of TAp63γ mutants on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression. p63, a member of the p53 gene family, known to play a role in development, has more recently also been implicated in cancer progression. Mice lacking p63 exhibit severe developmental defects such as limb truncations, abnormal skin, and absence of hair follicles, teeth, and mammary glands. Germline missense mutations of p63 have been shown to be responsible for several human developmental syndromes including SHFM, EEC and ADULT syndromes and are associated with anomalies in the development of organs of epithelial origin. -
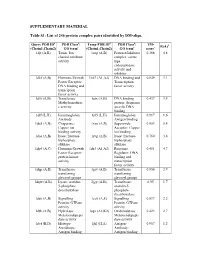
List of 246 Protein Complex Pairs Identified by DM-Align
SUPPLEMENTARY MATERIAL Table S1: List of 246 protein complex pairs identified by DM-align. Query PDB IDa PDB Classb: Temp PDB IDd PDB Classb: TM- R(Å)f (Chain1,Chain2) GO termc (Chain1,Chain2) GO termc scoree 1djt (A,B) Toxin: Ion 1aap (A,B) Protease/Inhibitor 0.368 4.8 channel inhibitor complex: serine activity type endopeptidase activity and inhibitor 1dkf (A,B) Hormone/Growth 1xb7 (A1,A2) DNA binding and 0.849 3.1 Factor Receptor: Transcription DNA binding and factor activity transciption factor activity 1dl5 (A,B) Transferase: 1utx (A,B) DNA binding 0.437 3.9 Methyltransferas protein: Sequence e activity specific DNA binding 1dlf (L,H) Immunoglobin: 1j05 (L,H) Immunoglobin: 0.917 1.6 Antibody Antigen binding 1do5 (A,B) Chaperone: 1xso (A,B) Superoxide 0.953 0.9 Copper ion Acceptor: Copper binding activity ion binding 1dos (A,B) lyase: fructose- 1rvg (A,B) lyase: fructose- 0.760 3.6 biphosphate biphosphate aldolase aldolase 1dp4 (A,C) Hormone/Growth 1dz3 (A1,A2) Response 0.481 4.7 Factor Receptor: Regulator: DNA protein kinase binding and activity transcription factor activity 1dqp (A,B) Transferase: 1grv (A,B) Transferase: 0.856 2.9 transferring transferring glycosyl groups glycosyl groups 1dqw (A,B) Lyase: orotidin- 2jgy (A,B) Transferase: 0.95 1.7 5-phosphate orotidin-5- decarboxylase phosphate decarboxylase 1ds6 (A,B) Signalling 1cc0 (A,E) Signalling 0.897 2.2 Protein: GTPase Protein: GTPase activity activity 1dth (A,B) Hydrolase: 1sqv (A2,K2) Oxidoredutase: 0.423 2.7 Metaloendopepti Metaloendopepti dase activity dase activity 1dvf -

Using MALDI-TOF MS Analysis for Rapid Discrimination of MRSA MLST
Using MALDI-TOF MS analysis for rapid discrimination of MRSA MLST types Jang-Jih Lu1,2,Tsui-Ping Liu1, Shih-Cheng Chang1,2 1Department of Laboratory Medicine, Chang Gung Memorial Hospital, Linkou, Taoyuan, Taiwan 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan Email: [email protected] 73 0:F8 MS Ra w 71 0:F9 MS Ra w 5000 6422.794 6000 Intens.[a.u.] Intens.[a.u.] Introduction 4000 Results ST5 ST5 4000 3000 2978.985 2000 6551.531 Staphylococcus aureus is one of the common causes of 3027.802 3054.785 2964.900 3175.943 2000 Table 1: MRSA and MSSA associated peaks by MALDI TOF 1000 community-associated infections and health care related 234 0:F1 MS Ra w 234 0:F1 MS Ra w 3005.919 MRSA(129 isolates) MSSA(77 isolates) 5000 6000 Intens.[a.u.] Intens.[a.u.] pathogen. It can usually cause wound infection, ST45 4000 ST45 6549.206 4000 3000 MS peaks (m/z) no. of isolate % of isolate no. of isolate % of isolate 6420.525 osteomyelitis, and even serious invasive infections, such as 2000 6610.628 2000 2978.373 3053.663 3174.805 2415 67 51.9 2 2.6 1000 sepsis and pneumonia. Epidemiology studies have shown 86 0:E4 MS Ra w 70 0:E5 MS Ra w 3058.565 5000 2431 65 50.4 0 0 3074.390 6000 Intens.[a.u.] Intens.[a.u.] that there are close relationship between the results of ST59 4000 6550.723 ST59 4000 3000 2879 51 39.5 2 2.6 6422.151 genotyping from multilocus sequence typing (MLST) and 2000 2000 characteristics of MRSA strains, including hospital- 6593 77 59.7 7 9.1 1000 200 0:E8 MS Ra w 38 0:E11 MS Ra w Appear of any above 103 79.8 11 14.3 2978.890 5000 6000 Intens.[a.u.] Intens.[a.u.] 6421.658 acquired, community-associated strain and even the 2965.568 ST239 4000 ST239 peaks 4000 3000 none of above peaks 6590.270 26 20.1 66 85.7 2937.113 3177.333 2916.384 antibiogram of the strain. -

HOXB6 Homeo Box B6 HOXB5 Homeo Box B5 WNT5A Wingless-Type
5 6 6 5 . 4 2 1 1 1 2 4 6 4 3 2 9 9 7 0 5 7 5 8 6 4 0 8 2 3 1 8 3 7 1 0 0 4 0 2 5 0 8 7 5 4 1 1 0 3 6 0 4 8 3 7 4 7 6 9 6 7 1 5 0 8 1 4 1 1 7 1 0 0 4 2 0 8 1 1 1 2 5 3 5 0 7 2 6 9 1 2 1 8 3 5 2 9 8 0 6 0 9 5 1 9 9 2 1 1 6 0 2 3 0 3 6 9 1 6 5 5 7 1 1 2 1 1 7 5 4 6 6 4 1 1 2 8 4 7 1 6 2 7 7 5 4 3 2 4 3 6 9 4 1 7 1 3 4 1 2 1 3 1 1 4 7 3 1 1 1 1 5 3 2 6 1 5 1 3 5 4 5 2 3 1 1 6 1 7 3 2 5 4 3 1 6 1 5 3 1 7 6 5 1 1 1 4 6 1 6 2 7 2 1 2 e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S HOXB6 homeo box B6 HOXB5 homeo box B5 WNT5A wingless-type MMTV integration site family, member 5A WNT5A wingless-type MMTV integration site family, member 5A FKBP11 FK506 binding protein 11, 19 kDa EPOR erythropoietin receptor SLC5A6 solute carrier family 5 sodium-dependent vitamin transporter, member 6 SLC5A6 solute carrier family 5 sodium-dependent vitamin transporter, member 6 RAD52 RAD52 homolog S. -

THE ALPHA-GALACTOSIDASE SUPERFAMILY: SEQUENCE BASED CLASSIFICATION of ALPHA-GALACTOSIDASES and RELATED GLYCOSIDASES Naumoff D.G
COMPUTATIONAL STRUCTURAL AND FUNCTIONAL PROTEOMICS THE ALPHA-GALACTOSIDASE SUPERFAMILY: SEQUENCE BASED CLASSIFICATION OF ALPHA-GALACTOSIDASES AND RELATED GLYCOSIDASES Naumoff D.G. State Institute for Genetics and Selection of Industrial Microorganisms, Moscow, Russia, e-mail: [email protected] Keywords: α-galactosidase, melibiase, glycoside hydrolase, GH-D clan, GH31 family, GHX family, COG1649, enzyme classification, protein family, protein phylogeny Summary Motivation: About 1 % of genes in genomes code enzymes with glycosidase activities. On the basis of sequence similarity all known glycosidases have been classified into 90 families. In many cases proteins of different families have common evolution origin. It makes necessary to combine the corresponding families into a superfamily. Results: Using of the PSI-BLAST program we found significant sequence similarity of several glycosidase families, two of which includes enzymes with the α galactosidase activity. Sequence homology, common catalytic mechanism, folding similarities, and composition of the active center allowed us to group three of these families – GH27, GH31, and GH36 – into the α-galactosidase superfamily. Phylogenetic analysis of this superfamily revealed polyphyletic origin of GH36 family, which could be divided into four families. Glycosidases of the α-galactosidase superfamily have a distant relationship with proteins belonging to families GH13, GH70, and GH77 of glycosidases, as well as with two families of predicted glycosidases. Introduction Glycoside hydrolases or glycosidases (EC 3.2.1.-) are a widespread group of enzymes, hydrolyzing the glycosidic bonds between two carbohydrates or between a carbohydrate and an aglycone moiety. A large multiplicity of these enzymes is a consequence of the extensive variety of their natural substrates: di-, oligo-, and polysaccharides. -

(12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown Et Al
US 20030082511A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown et al. (43) Pub. Date: May 1, 2003 (54) IDENTIFICATION OF MODULATORY Publication Classification MOLECULES USING INDUCIBLE PROMOTERS (51) Int. Cl." ............................... C12O 1/00; C12O 1/68 (52) U.S. Cl. ..................................................... 435/4; 435/6 (76) Inventors: Steven J. Brown, San Diego, CA (US); Damien J. Dunnington, San Diego, CA (US); Imran Clark, San Diego, CA (57) ABSTRACT (US) Correspondence Address: Methods for identifying an ion channel modulator, a target David B. Waller & Associates membrane receptor modulator molecule, and other modula 5677 Oberlin Drive tory molecules are disclosed, as well as cells and vectors for Suit 214 use in those methods. A polynucleotide encoding target is San Diego, CA 92121 (US) provided in a cell under control of an inducible promoter, and candidate modulatory molecules are contacted with the (21) Appl. No.: 09/965,201 cell after induction of the promoter to ascertain whether a change in a measurable physiological parameter occurs as a (22) Filed: Sep. 25, 2001 result of the candidate modulatory molecule. Patent Application Publication May 1, 2003 Sheet 1 of 8 US 2003/0082511 A1 KCNC1 cDNA F.G. 1 Patent Application Publication May 1, 2003 Sheet 2 of 8 US 2003/0082511 A1 49 - -9 G C EH H EH N t R M h so as se W M M MP N FIG.2 Patent Application Publication May 1, 2003 Sheet 3 of 8 US 2003/0082511 A1 FG. 3 Patent Application Publication May 1, 2003 Sheet 4 of 8 US 2003/0082511 A1 KCNC1 ITREXCHO KC 150 mM KC 2000000 so 100 mM induced Uninduced Steady state O 100 200 300 400 500 600 700 Time (seconds) FIG. -

In TCR-Stimulated Thymocytes Induction of Chromosomal DNA Degradation Caspase-Activated Deoxyribonuclease in the Possible Involv
Possible Involvement of Cyclophilin B and Caspase-Activated Deoxyribonuclease in the Induction of Chromosomal DNA Degradation in TCR-Stimulated Thymocytes This information is current as of September 28, 2021. Takuya Nagata, Hiroyuki Kishi, Qing Li Liu, Tomoyasu Yoshino, Tadashi Matsuda, Zhe Xiong Jin, Kimie Murayama, Kazuhiro Tsukada and Atsushi Muraguchi J Immunol 2000; 165:4281-4289; ; doi: 10.4049/jimmunol.165.8.4281 Downloaded from http://www.jimmunol.org/content/165/8/4281 References This article cites 42 articles, 23 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/165/8/4281.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 28, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2000 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Possible Involvement of Cyclophilin B and Caspase-Activated Deoxyribonuclease in the Induction of Chromosomal DNA Degradation in TCR-Stimulated Thymocytes1 Takuya Nagata,* Hiroyuki Kishi,* Qing Li Liu,* Tomoyasu Yoshino,* Tadashi Matsuda,* Zhe Xiong Jin,* Kimie Murayama,‡ Kazuhiro Tsukada,† and Atsushi Muraguchi2* TCR engagement of immature CD4؉CD8؉ thymocytes induces clonal maturation (positive selection) as well as clonal deletion (negative selection) in the thymus. -

Curcumin Alters Gene Expression-Associated DNA Damage, Cell Cycle, Cell Survival and Cell Migration and Invasion in NCI-H460 Human Lung Cancer Cells in Vitro
ONCOLOGY REPORTS 34: 1853-1874, 2015 Curcumin alters gene expression-associated DNA damage, cell cycle, cell survival and cell migration and invasion in NCI-H460 human lung cancer cells in vitro I-TSANG CHIANG1,2, WEI-SHU WANG3, HSIN-CHUNG LIU4, SU-TSO YANG5, NOU-YING TANG6 and JING-GUNG CHUNG4,7 1Department of Radiation Oncology, National Yang‑Ming University Hospital, Yilan 260; 2Department of Radiological Technology, Central Taiwan University of Science and Technology, Taichung 40601; 3Department of Internal Medicine, National Yang‑Ming University Hospital, Yilan 260; 4Department of Biological Science and Technology, China Medical University, Taichung 404; 5Department of Radiology, China Medical University Hospital, Taichung 404; 6Graduate Institute of Chinese Medicine, China Medical University, Taichung 404; 7Department of Biotechnology, Asia University, Taichung 404, Taiwan, R.O.C. Received March 31, 2015; Accepted June 26, 2015 DOI: 10.3892/or.2015.4159 Abstract. Lung cancer is the most common cause of cancer CARD6, ID1 and ID2 genes, associated with cell survival and mortality and new cases are on the increase worldwide. the BRMS1L, associated with cell migration and invasion. However, the treatment of lung cancer remains unsatisfactory. Additionally, 59 downregulated genes exhibited a >4-fold Curcumin has been shown to induce cell death in many human change, including the DDIT3 gene, associated with DNA cancer cells, including human lung cancer cells. However, the damage; while 97 genes had a >3- to 4-fold change including the effects of curcumin on genetic mechanisms associated with DDIT4 gene, associated with DNA damage; the CCPG1 gene, these actions remain unclear. Curcumin (2 µM) was added associated with cell cycle and 321 genes with a >2- to 3-fold to NCI-H460 human lung cancer cells and the cells were including the GADD45A and CGREF1 genes, associated with incubated for 24 h.