In TCR-Stimulated Thymocytes Induction of Chromosomal DNA Degradation Caspase-Activated Deoxyribonuclease in the Possible Involv
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown Et Al
US 20030082511A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown et al. (43) Pub. Date: May 1, 2003 (54) IDENTIFICATION OF MODULATORY Publication Classification MOLECULES USING INDUCIBLE PROMOTERS (51) Int. Cl." ............................... C12O 1/00; C12O 1/68 (52) U.S. Cl. ..................................................... 435/4; 435/6 (76) Inventors: Steven J. Brown, San Diego, CA (US); Damien J. Dunnington, San Diego, CA (US); Imran Clark, San Diego, CA (57) ABSTRACT (US) Correspondence Address: Methods for identifying an ion channel modulator, a target David B. Waller & Associates membrane receptor modulator molecule, and other modula 5677 Oberlin Drive tory molecules are disclosed, as well as cells and vectors for Suit 214 use in those methods. A polynucleotide encoding target is San Diego, CA 92121 (US) provided in a cell under control of an inducible promoter, and candidate modulatory molecules are contacted with the (21) Appl. No.: 09/965,201 cell after induction of the promoter to ascertain whether a change in a measurable physiological parameter occurs as a (22) Filed: Sep. 25, 2001 result of the candidate modulatory molecule. Patent Application Publication May 1, 2003 Sheet 1 of 8 US 2003/0082511 A1 KCNC1 cDNA F.G. 1 Patent Application Publication May 1, 2003 Sheet 2 of 8 US 2003/0082511 A1 49 - -9 G C EH H EH N t R M h so as se W M M MP N FIG.2 Patent Application Publication May 1, 2003 Sheet 3 of 8 US 2003/0082511 A1 FG. 3 Patent Application Publication May 1, 2003 Sheet 4 of 8 US 2003/0082511 A1 KCNC1 ITREXCHO KC 150 mM KC 2000000 so 100 mM induced Uninduced Steady state O 100 200 300 400 500 600 700 Time (seconds) FIG. -

Curcumin Alters Gene Expression-Associated DNA Damage, Cell Cycle, Cell Survival and Cell Migration and Invasion in NCI-H460 Human Lung Cancer Cells in Vitro
ONCOLOGY REPORTS 34: 1853-1874, 2015 Curcumin alters gene expression-associated DNA damage, cell cycle, cell survival and cell migration and invasion in NCI-H460 human lung cancer cells in vitro I-TSANG CHIANG1,2, WEI-SHU WANG3, HSIN-CHUNG LIU4, SU-TSO YANG5, NOU-YING TANG6 and JING-GUNG CHUNG4,7 1Department of Radiation Oncology, National Yang‑Ming University Hospital, Yilan 260; 2Department of Radiological Technology, Central Taiwan University of Science and Technology, Taichung 40601; 3Department of Internal Medicine, National Yang‑Ming University Hospital, Yilan 260; 4Department of Biological Science and Technology, China Medical University, Taichung 404; 5Department of Radiology, China Medical University Hospital, Taichung 404; 6Graduate Institute of Chinese Medicine, China Medical University, Taichung 404; 7Department of Biotechnology, Asia University, Taichung 404, Taiwan, R.O.C. Received March 31, 2015; Accepted June 26, 2015 DOI: 10.3892/or.2015.4159 Abstract. Lung cancer is the most common cause of cancer CARD6, ID1 and ID2 genes, associated with cell survival and mortality and new cases are on the increase worldwide. the BRMS1L, associated with cell migration and invasion. However, the treatment of lung cancer remains unsatisfactory. Additionally, 59 downregulated genes exhibited a >4-fold Curcumin has been shown to induce cell death in many human change, including the DDIT3 gene, associated with DNA cancer cells, including human lung cancer cells. However, the damage; while 97 genes had a >3- to 4-fold change including the effects of curcumin on genetic mechanisms associated with DDIT4 gene, associated with DNA damage; the CCPG1 gene, these actions remain unclear. Curcumin (2 µM) was added associated with cell cycle and 321 genes with a >2- to 3-fold to NCI-H460 human lung cancer cells and the cells were including the GADD45A and CGREF1 genes, associated with incubated for 24 h. -

Cleavage and Nuclear Translocation of the Caspase 3 Substrate Rho GDP-Dissociation Inhibitor, D4-GDI, During Apoptosis
Cell Death and Differentiation (1999) 6, 412 ± 419 ã 1999 Stockton Press All rights reserved 13509047/99 $12.00 http://www.stockton-press.co.uk/cdd Cleavage and nuclear translocation of the caspase 3 substrate Rho GDP-dissociation inhibitor, D4-GDI, during apoptosis 1 ,1 Ronald J Krieser and Alan Eastman* Introduction 1 Department of Pharmacology and Toxicology, Dartmouth Medical School, Apoptosis plays a central role in such processes as Hanover, New Hampshire 03755, USA development, tissue homeostasis, and thymic selection, as * corresponding author: tel: (603)-650-1501; fax: (603)-650-1129; well as pathologies ranging from neurodegenerative disease, e-mail: [email protected] autoimmune disorders, and viral infection, to cancer. Much research on apoptosis has focused on determining proteins involved in decisions of cell fate, and regulation of the Received 17.11.98; revised 15.2.99; accepted 2.3.99 execution phase of cell death involving protease and Edited by G. Salvesen endonuclease activation. A great deal of this information has been gained from the study of small organisms such as Abstract the nematode C. elegans, which has a well-defined developmental program during which specific cells die. The While investigating endonucleases potentially involved in characterization of nematodes with mutations in the cell death apoptosis, an antisera was raised to bovine deoxyribonu- process has led to identification of genes which regulate clease II, but it recognized a smaller protein of 26 kDa protein apoptosis.1 The mammalian homologs have been identified in a variety of cell lines. The 26 kDa protein underwent for many of these regulatory genes. -

Identification of Oxidative Stress-Related Proteins for Predictive Screening of Hepatotoxicity Using a Proteomic Approach
The Journal of Toxicological Sciences, 213 Vol.30, No.3, 213-227, 2005 IDENTIFICATION OF OXIDATIVE STRESS-RELATED PROTEINS FOR PREDICTIVE SCREENING OF HEPATOTOXICITY USING A PROTEOMIC APPROACH Toshinori YAMAMOTO, Rie KIKKAWA, Hiroshi YAMADA and Ikuo HORII Worldwide Safety Sciences, Pfizer Global Research & Development, Nagoya Laboratories, Pfizer Inc., 5-2 Taketoyo, Aichi 470-2393, Japan (Received January 15, 2005; Accepted April 19, 2005) ABSTRACT — We investigated the effects of three hepatotoxicants, acetaminophen (APAP), amio- darone (AD) and tetracycline (TC), on protein expression in primary cultured rat hepatocytes with toxi- coproteomic approach, which is two-dimensional gel electrophoresis (2DE) and mass spectrometry. The objectives of this study were to search for alternative toxicity biomarkers which could be detected with high sensitivity prior to the appearance of morphological changes or alterations of analytical conventional biomarkers. The related proteins in the process of cell degeneration/necrosis such as cell death, lipid metabolism and lipid/carbohydrate metabolism were mainly affected under exposure to APAP, AD and TC, respectively. Among the differentially expressed proteins, several oxidative stress-related proteins were clearly identified after 24-hr exposure, even though they were not affected for 6-hr exposure. They were glutathione peroxidase (GPX) as a down-regulated protein as well as peroxiredoxin 1 (PRX1) and peroxiredoxin 2 (PRX2) as up-regulated proteins, which are known to serve as antioxidative enzymes in cells. These findings suggested that the focused proteins, GPX and PRXs, could be utilized as biomarkers of hepatotoxicity, and they were useful for setting high throughput screening methods to assess hepato- toxicity in the early stage of drug discovery. -

Supplementary Material Toxicological Impacts and Likely Protein Targets Of
Supplementary Material Toxicological impacts and likely protein targets of bisphenol A in Paramecium caudatum Marcus V. X. Senra† & Ana Lúcia Fonseca Instituto de Recursos Naturais, Universidade Federal de Itajubá, 37500-903, Itajubá, Minas Gerais – Brazil †To whom correspondence should be addressed – [email protected]; Orcid - 0000-0002-3866- 8837 Table S1. Annotation data on the P. caudatum 3D modelled proteins and their binding energies to BPA. BINDING ID DESCRIPTION CHROMOSOME NT_START NT_END ENERGIES (kcal/mol) PCAU.43c3d.1.P00010012 Tryptophan synthase beta subunit-like PLP-dependent enzyme scaffold_0001 23197 24853 -7.4 PCAU.43c3d.1.P00010015 Ribosomal protein L32e scaffold_0001 26373 26859 -6.2 PCAU.43c3d.1.P00010044 Catalase, mono-functional, haem-containing scaffold_0001 71821 73367 -6.5 PCAU.43c3d.1.P00010050 Dihydroorotate dehydrogenase, class 1/ 2 scaffold_0001 76614 79650 -6.6 PCAU.43c3d.1.P00010054 Serine/threonine/dual specificity protein kinase, catalytic domain scaffold_0001 83399 84653 -6.7 PCAU.43c3d.1.P00010070 Peptidyl-prolyl cis-trans isomerase, FKBP-type scaffold_0001 104909 105387 -5.9 PCAU.43c3d.1.P00010103 V-ATPase proteolipid subunit C-like domain scaffold_0001 168736 169346 -5.6 PCAU.43c3d.1.P00010112 DNA-directed RNA polymerase, RBP11-like dimerisation domain scaffold_0001 180310 181372 -6.0 PCAU.43c3d.1.P00010165 Vacuolar (H+)-ATPase G subunit scaffold_0001 252653 253112 -5.6 PCAU.43c3d.1.P00010176 Coproporphyrinogen III oxidase, aerobic scaffold_0001 262051 263168 -6.7 PCAU.43c3d.1.P00010205 Metalloenzyme, -

However, There Was Substantial Falloff in Terms Of
446 Technical Briefs accuracy; however, there was substantial falloff in terms disease, myocardial infarction, and ischemic cerebrovascular disease. Six case-control studies from the Copenhagen City Heart Study. Ann Intern Med of performance with new vs used chips. This lowers the 2001;134:941–54. cost per SNP to almost one-third of the cost of using a new 5. Twyman RM, Primrose SB. Techniques patents for SNP genotyping. Pharma- microelectronic chip and more lowers the cost compared cogenomics 2003;4:67–79. 6. Thistlethwaite WA. Rapid genotyping of common MeCP2 mutations with an with RFLP analysis by more than one-half. This is approx- electronic DNA microchip using serial differential hybridization. J Mol Diagn imately the same cost reported by others for detecting 2003;5:121–6. eight SNPs on one test site simultaneously (€1.62 per SNP) 7. Gilles PN, Wu DJ, Foster CB, Dillon PJ, Chanock SJ. Single nucleotide polymorphic discrimination by an electronic dot blot assay on semiconductor (6). It should be noted, however, that purchase of the microchips. Nat Biotechnol 1999;17:365–70. Nanogen NMW 1000 Nanochip Molecular Biology Work- 8. Nagan N, O’Kane DJ. Validation of a single nucleotide polymorphism station is not included in these calculations. genotyping assay for the human serum paraoxonase gene using electroni- cally active customized microarrays. Clin Biochem 2001;34:589–92. The microelectronic chip examined in this study was 9. Sohni YR, Dukek B, Taylor W, Ricart E, Sandborn WJ, O’Kane DJ. Active limited by the number of test sites per chip. Increasing the electronic arrays for genotyping of NAT2 polymorphisms. -

Papadaki Et Al., 2009 Supplementary
Papadaki et al., 2009 Supplementary Supplemental Data Index x Supplemental Figures 1-6 x Supplemental Tables 1a, 1b, 2 Papadaki et al., 2009 Supplementary Supplemental Figure 1. Thymocyte restricted inactivation of the Elavl1 locus. + fl (A) Diagrammatic representation of the wild-type (Elavl1P P), floxed (Elavl1P P) and Cre- - recombined (Elavl1P P) Elavl1/HuR loci on mouse chromosome 8; Noted are the loxP sequences (triangles) flanking the selection marker (neo) used in gene targeting and the ATG containing exon 2 (white box); (H) denotes restriction sites for loci mapping. (B) Detection of native (+), targeted (fl) and Cre-recombinant (-) loci in thymocyte DNA extracts from control and test mice following HindIII digestion and Southern blotting. (C) Western blot of total thymic protein extracts probed with ĮHuR Ab + fl/fl indicating the loss of HuR protein in LckCreP PElavl1P P thymi. Į-actin is shown for quantitation. (D) Flow cytometric detection of intracellular mHuR protein in + fl/+ LckCreP PElavl1P P thymocytes (open histogram), and its respective loss in + fl/fl LckCreP PElavl1P P thymocytes (shaded histogram). The dotted histogram depicts the + isotype-matched background staining. (E) Flow cytometric detection of HuRP P or - + + + fl/+ HuRP P cells in gated splenic CD4P Por CD8P P T-cells from 8 week old LckCreP PElavl1P + fl/fl - Pand LckCreP PElavl1P P mice respectively. (F) Enumeration of HuRP P cells in + fl/fl LckCreP PElavl1P P thymocyte subsets and splenic T-cells; Data are percentages (+SEM) derived from the flow cytometric detection of HuR- cells in CD4/CD8/DP and DN gated populations (n=12-15) at 8-10 weeks of age. -

Identification of Genomic Targets of Krüppel-Like Factor 9 in Mouse Hippocampal
Identification of Genomic Targets of Krüppel-like Factor 9 in Mouse Hippocampal Neurons: Evidence for a role in modulating peripheral circadian clocks by Joseph R. Knoedler A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Neuroscience) in the University of Michigan 2016 Doctoral Committee: Professor Robert J. Denver, Chair Professor Daniel Goldman Professor Diane Robins Professor Audrey Seasholtz Associate Professor Bing Ye ©Joseph R. Knoedler All Rights Reserved 2016 To my parents, who never once questioned my decision to become the other kind of doctor, And to Lucy, who has pushed me to be a better person from day one. ii Acknowledgements I have a huge number of people to thank for having made it to this point, so in no particular order: -I would like to thank my adviser, Dr. Robert J. Denver, for his guidance, encouragement, and patience over the last seven years; his mentorship has been indispensable for my growth as a scientist -I would also like to thank my committee members, Drs. Audrey Seasholtz, Dan Goldman, Diane Robins and Bing Ye, for their constructive feedback and their willingness to meet in a frequently cold, windowless room across campus from where they work -I am hugely indebted to Pia Bagamasbad and Yasuhiro Kyono for teaching me almost everything I know about molecular biology and bioinformatics, and to Arasakumar Subramani for his tireless work during the home stretch to my dissertation -I am grateful for the Neuroscience Program leadership and staff, in particular -

Proteomics of the Lysosome
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biochimica et Biophysica Acta 1793 (2009) 625–635 Contents lists available at ScienceDirect Biochimica et Biophysica Acta journal homepage: www.elsevier.com/locate/bbamcr Review Proteomics of the lysosome Torben Lübke a, Peter Lobel b,c, David E. Sleat b,c,⁎ a Zentrum Biochemie und Molekulare Zellbiologie, Abteilung Biochemie II, Georg-August Universität Göttingen, 37073 Göttingen, Germany b Center for Advanced Biotechnology and Medicine, Piscataway, NJ 08854, USA c Department of Pharmacology, University of Medicine and Dentistry of New Jersey - Robert Wood Johnson Medical School, Piscataway, NJ 08854, USA article info abstract Article history: Defects in lysosomal function have been associated with numerous monogenic human diseases typically Received 16 May 2008 classified as lysosomal storage diseases. However, there is increasing evidence that lysosomal proteins are Received in revised form 24 September 2008 also involved in more widespread human diseases including cancer and Alzheimer disease. Thus, there is a Accepted 30 September 2008 continuing interest in understanding the cellular functions of the lysosome and an emerging approach to this Available online 15 October 2008 is the identification of its constituent proteins by proteomic analyses. To date, the mammalian lysosome has been shown to contain ∼60 soluble luminal proteins and ∼25 transmembrane proteins. However, recent Keywords: fi fi Lysosomal protein proteomic studies based upon af nity puri cation of soluble components or subcellular fractionation to Proteomic obtain both soluble and membrane components suggest that there may be many more of both classes of Mass spectrometry protein resident within this organelle than previously appreciated. -
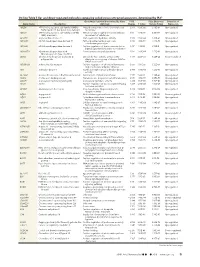
Up- and Down-Regulated Molecules Comparing Coiled Versus
On-line Table 1: Up- and down-regulated molecules comparing coiled versus untreated aneurysms, determined by IPAa Gene Main Function Determined by Gene Fold False Discovery Direction of Gene Name Description Ontology Change P Value Rate (q Value) Expression ABCB6 ATP-binding cassette, sub-family B (MDR/ Integral component of mitochondrial outer 2.541 4.28E-05 5.90E-04 Up-regulated TAP), member 6 (Langereis blood group) membrane ABCC1 ATP-binding cassette, sub-family C (CFTR/ ATPase activity, coupled to transmembrane 3.132 4.94E-10 6.17E-08 Up-regulated MRP), member 1 movement of substances ACOT11 Acyl-coa thioesterase 11 Carboxylic ester hydrolase activity 3.973 2.58E-04 2.42E-03 Up-regulated ADAM10 ADAM metallopeptidase domain 10 PMA-inducible membrane protein 3.425 1.33E-04 1.44E-03 Up-regulated ectodomain proteolysis ADAM8 ADAM metallopeptidase domain 8 Positive regulation of tumor necrosis factor 3.417 1.42E-13 6.74E-11 Up-regulated (ligand) superfamily member 11 production ADAMTS4 ADAM metallopeptidase with Proteinaceous extracellular matrix 2.134 2.49E-08 1.52E-06 Up-regulated thrombospondin type 1 motif, 4 ADH4 Alcohol dehydrogenase 4 (class II), pi Oxidoreductase activity, acting on the Ϫ2.657 4.44E-05 6.04E-04 Down-regulated polypeptide aldehyde or oxo group of donors, NAD or NADP as acceptor ADORA2B Adenosine A2b receptor Positive regulation of chronic inflammatory 2.056 1.95E-05 3.22E-04 Up-regulated response to non-antigenic stimulus AK4 Adenylate kinase 4 Nucleoside triphosphate adenylate kinase 2.074 1.96E-08 1.25E-06 Up-regulated -

Disruption of the Gene in a Mouse Model
The Role of Dl,lase X in Skeletal Muscle Addressed by Targeted Disruption of the Gene in a Mouse Model By Iran Røshedi A thesis submitted to The Faculty of Graduate Studies of the University of Manitoba ln partial fulfillment of the requirements for the degree of Masterts of Science Department of Biochemistry and Medical Genetics Faculty of Medicine University of Manitoba Winnipeg Copyright O 2008 by Iran Rashedi THE I]NIVERSITY OF MANITOBA F'ÄCULTY OF GRADUÄTE STI]DIES t<¡t*t<* COPYRIGHT PERMISSION The Role of DNase X in Skeletal Muscle Addressed by Targeted Disruption of the Gene in a Mouse Model BY Iran Rashedi A ThesislPracticum submitted to the Faculty of Graduate Studies of The University of Manitoba in partial fulfillment of the requirement of the degree of MASTER OF SCIENCE Iran Rashedi O 2008 Permission has been granted to the University of Manitoba Libraries to lend a copy of this thesis/practicum, to Library and Archives Canada (tAC) to lend a copy of this thesis/practicum, and to LAC's agent (UMlÆroQuest) to microfilm, sell copies and to publish an abstract of this thesis/practicum. This reproduction or copy of this thesis has been made available by authority of the copyright owner solely for the purpose of private study and research, and may only be reproduced and copied as permitted by copyright laws or with express written authorization from the copyright owner. This work is dedícated to my deørly lovedfømily whose ever-løsting support gives me the chønce to follow my dreams, ønd to øll my teøchers who høve tøught me greøt lessons both in my lífe ønd my øcødemíc cøreer. -

Supplemental Data
Supplementary Table 1. Gene sets from Figure 6. Lists of genes from each individual gene set defined in Figure 6, including the fold-change in expression of each gene in treatment group pair-wise comparisons. ENSEMBL: Ensembl gene identifier; Symbol: official gene symbol; logFC: log fold change; p value: significance of fold-change in a pair-wise comparison, P<0.05 cut-off; FDR: false discovery rate, expected proportion of false positives among the differentially expressed genes in a pair-wise comparison (FDR<0.25 cut-off). Sup. Table 1 SET I CP versus Sal CP versus CP+DCA DCA versus Sal ENSEMBL Symbol logFC PValue FDR logFC PValue FDR logFC PValue FDR Desc ENSMUSG00000020326 Ccng1 2.64 0.00 0.00 -0.06 0.13 0.96 0.40 0.00 0.23 cyclin G1 [Source:MGI Symbol;Acc:MGI:102890] ENSMUSG00000031886 Ces2e 3.97 0.00 0.00 -0.24 0.02 0.28 0.01 1.00 1.00 carboxylesterase 2E [Source:MGI Symbol;Acc:MGI:2443170] ENSMUSG00000041959 S100a10 2.31 0.00 0.00 -0.21 0.02 0.23 -0.11 0.53 1.00 S100 calcium binding protein A10 (calpactin) [Source:MGI Symbol;Acc:MGI:1339468] ENSMUSG00000092341 Malat1 1.09 0.00 0.00 -0.11 0.20 1.00 0.66 0.00 0.00 metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) [Source:MGI Symbol;Acc:MGI:1919539] ENSMUSG00000072949 Acot1 1.73 0.00 0.00 -0.22 0.01 0.12 -0.44 0.01 1.00 acyl-CoA thioesterase 1 [Source:MGI Symbol;Acc:MGI:1349396] ENSMUSG00000064339 mt-Rnr2 1.09 0.00 0.00 -0.08 0.17 1.00 0.67 0.00 0.07 mitochondrially encoded 16S rRNA [Source:MGI Symbol;Acc:MGI:102492] ENSMUSG00000025934 Gsta3 1.86 0.00 0.00 -0.28