Sigma Enzymes, Coenzymes and Enzyme Substrates
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Genome-Wide Transcriptional Changes and Lipid Profile
G C A T T A C G G C A T genes Article Genome-Wide Transcriptional Changes and Lipid Profile Modifications Induced by Medicago truncatula N5 Overexpression at an Early Stage of the Symbiotic Interaction with Sinorhizobium meliloti Chiara Santi 1, Barbara Molesini 1, Flavia Guzzo 1, Youry Pii 2 ID , Nicola Vitulo 1 and Tiziana Pandolfini 1,* ID 1 Department of Biotechnology, University of Verona, 37134 Verona, Italy; [email protected] (C.S.); [email protected] (B.M.); fl[email protected] (F.G.); [email protected] (N.V.) 2 Faculty of Science and Technology, Free University of Bozen-Bolzano, 39100 Bolzano BZ, Italy; [email protected] * Correspondence: tiziana.pandolfi[email protected]; Tel.: +39-045-8027918 Received: 30 October 2017; Accepted: 11 December 2017; Published: 19 December 2017 Abstract: Plant lipid-transfer proteins (LTPs) are small basic secreted proteins, which are characterized by lipid-binding capacity and are putatively involved in lipid trafficking. LTPs play a role in several biological processes, including the root nodule symbiosis. In this regard, the Medicago truncatula nodulin 5 (MtN5) LTP has been proved to positively regulate the nodulation capacity, controlling rhizobial infection and nodule primordia invasion. To better define the lipid transfer protein MtN5 function during the symbiosis, we produced MtN5-downregulated and -overexpressing plants, and we analysed the transcriptomic changes occurring in the roots at an early stage of Sinorhizobium meliloti infection. We also carried out the lipid profile analysis of wild type (WT) and MtN5-overexpressing roots after rhizobia infection. The downregulation of MtN5 increased the root hair curling, an early event of rhizobia infection, and concomitantly induced changes in the expression of defence-related genes. -

Proteomic Analysis of Mycelial Proteins from Magnaporthe Oryzae Under Nitrogen Starvation
Proteomic analysis of mycelial proteins from Magnaporthe oryzae under nitrogen starvation X.-G. Zhou1,2, P. Yu 2, C. Dong2, C.-X. Yao2, Y.-M. Ding2, N. Tao2 and Z.-W. Zhao1 1State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, Kunming, China 2Key Laboratory of Southwestern Crop Gene Resources and Germplasm Innovation, Ministry of Agriculture and Yunnan Provincial Key Laboratory of Agricultural Biotechnology, Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences, Kunming, China Corresponding author: Z.W. Zhao E-mail: [email protected] Genet. Mol. Res. 15 (2): gmr.15028637 Received March 23, 2016 Accepted April 11, 2016 Published May 13, 2016 DOI http://dx.doi.org/10.4238/gmr.15028637 ABSTRACT. Magnaporthe oryzae is an important model system in studies of plant pathogenic fungi, and nitrogen is a key nutrient source affecting microbial growth and development. In order to understand how nitrogen stress causes changes in mycelial proteins, we analyzed differentially expressed mycelial proteins from the M. oryzae virulent strain CH-63 using two-dimensional electrophoresis and mass spectrometry in complete medium or under nitrogen starvation conditions. A total of 975 ± 70 and 1169 ± 90 protein spots were detected in complete medium and under nitrogen starvation conditions, respectively. Forty-nine protein spots exhibited at least 2-fold up- regulation or down-regulation at the protein level according to PDQuest7.4. Moreover, 43 protein spots were successfully identified by matrix-assisted laser desorption/ionization-time-of-flight/time-of-flight mass spectrometry. Among these spots, 6 proteins were functionally unknown and 37 proteins were categorized into 5 groups according to Genetics and Molecular Research 15 (2): gmr.15028637 ©FUNPEC-RP www.funpecrp.com.br X.-G. -

Updating the Sequence-Based Classification of Glycosyl Hydrolases
Article Updating the sequence-based classification of glycosyl hydrolases HENRISSAT, Bernard, BAIROCH, Amos Marc Reference HENRISSAT, Bernard, BAIROCH, Amos Marc. Updating the sequence-based classification of glycosyl hydrolases. Biochemical Journal, 1996, vol. 316 ( Pt 2), p. 695-6 PMID : 8687420 DOI : 10.1042/bj3160695 Available at: http://archive-ouverte.unige.ch/unige:36909 Disclaimer: layout of this document may differ from the published version. 1 / 1 Biochem. J. (1996) 316, 695–696 (Printed in Great Britain) 695 BIOCHEMICAL JOURNAL Updating the sequence-based classification of available. When the number of glycosyl hydrolase sequences reached C 480, ten additional families (designated 36–45) could glycosyl hydrolases be defined and were added to the classification [2]. There are at present over 950 sequences of glycosyl hydrolases in the data- A classification of glycosyl hydrolases based on amino-acid- banks (EMBL}GenBank and SWISS-PROT). Their analysis sequence similarities was proposed in this Journal a few years shows that the vast majority of the C 470 additional sequences ago [1]. This classification originated from the analysis of C 300 that have become available since the last update could be classified sequences and their grouping into 35 families designated 1–35. in the existing families. However, several sequences not fitting Because such a classification is necessarily sensitive to the sample, the existing families allow the definition of new families (desig- it was anticipated that it was incomplete and that new families nated 46–57) (Table 1). When the several present genome would be determined when additional sequences would become sequencing projects have reached completion, the number of Table 1 New families in the classification of glycosyl hydrolases Family Enzyme Organism SWISS-PROT EMBL/GenBank 46 Chitosanase Bacillus circulans MH-K1 P33673 D10624 46 Chitosanase Streptomyces sp. -

Oxdc Antibody Rabbit Polyclonal Antibody Catalog # ABV11223
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 OxdC Antibody Rabbit Polyclonal Antibody Catalog # ABV11223 Specification OxdC Antibody - Product Information Application WB Primary Accession O34714 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit IgG Calculated MW 43566 OxdC Antibody - Additional Information Gene ID 938620 Positive Control Western Blot: Recombinant protein Application & Usage Western blot: 1-4 Western blot of Oxalate decarboxylase µg/ml. antibody. Lane 1: rb- Oxalate decarboxylase - Other Names 10 ng. Lane 2: rb- Oxalate decarboxylase - 50 YvrK ng Target/Specificity OxdC OxdC Antibody - Background Antibody Form Oxalate decarboxylase (OxdC, EC4.1.1.2) is a Liquid manganese-containing enzyme, which decomposes oxalic acid and oxalate. With Appearance OxdC catalysis, oxalate is split into formate Colorless liquid and CO2. This enzyme belongs to the family of lyases, specifically the carboxy-lyases, which Formulation 100 µg (0.5 mg/ml) of antibody in PBS pH cleave carbon-carbon bonds. The systematic 7.2 containing 0.01 % BSA, 0.01 % name of this enzyme class is oxalate thimerosal, and 50 % glycerol. carboxy-lyase (formate-forming). This enzyme is also called oxalate carboxy-lyase. The Handling enzyme is composed of two cupin domains, The antibody solution should be gently each of which contains a Mn (II) ion. This mixed before use. enzyme participates in glyoxylate and dicarboxylate metabolism. This enzyme has Reconstitution & Storage been recognized for diagnostics in diverse -20 °C biotechnological applications such as the clinical assay of oxalate in blood and urine, Background Descriptions therapeutics, process industry, and agriculture to lower oxalate levels in foods and the environment. -

Deletion of the Gene Encoding the Reductase Component of 3
Yeh et al. Microbial Cell Factories 2014, 13:130 http://www.microbialcellfactories.com/content/13/1/130 RESEARCH Open Access Deletion of the gene encoding the reductase component of 3-ketosteroid 9α-hydroxylase in Rhodococcus equi USA-18 disrupts sterol catabolism, leading to the accumulation of 3-oxo-23,24-bisnorchola-1,4-dien-22-oic acid and 1,4-androstadiene-3,17-dione Chin-Hsing Yeh1, Yung-Shun Kuo1, Che-Ming Chang1, Wen-Hsiung Liu2, Meei-Ling Sheu3 and Menghsiao Meng1* Abstract The gene encoding the putative reductase component (KshB) of 3-ketosteroid 9α-hydroxylase was cloned from Rhodococcus equi USA-18, a cholesterol oxidase-producing strain formerly named Arthrobacter simplex USA-18, by PCR according to consensus amino acid motifs of several bacterial KshB subunits. Deletion of the gene in R. equi USA-18 by a PCR-targeted gene disruption method resulted in a mutant strain that could accumulate up to 0.58 mg/ml 1,4-androstadiene-3,17-dione (ADD) in the culture medium when 0.2% cholesterol was used as the carbon source, indicating the involvement of the deleted enzyme in 9α-hydroxylation of steroids. In addition, this mutant also accumulated 3-oxo-23,24-bisnorchola-1,4-dien-22-oic acid (Δ1,4-BNC). Because both ADD and Δ1,4-BNC are important intermediates for the synthesis of steroid drugs, this mutant derived from R. equi USA-18 may deserve further investigation for its application potential. Background generally believed to be carried out by cholesterol oxidase, Steroid drugs, including androgens, anabolic steroids, es- which catalyzes the oxidation of the 3β-hydroxyl moiety of trogens and corticosteroids, have been widely used for a sterols with the simultaneous isomerization of Δ5 to Δ4, variety of health purposes. -

To Study Mutant P53 Gain of Function, Various Tumor-Derived P53 Mutants
Differential effects of mutant TAp63γ on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression. A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By Shama K Khokhar M.Sc., Bilaspur University, 2004 B.Sc., Bhopal University, 2002 2007 1 COPYRIGHT SHAMA K KHOKHAR 2007 2 WRIGHT STATE UNIVERSITY SCHOOL OF GRADUATE STUDIES Date of Defense: 12-03-07 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY SHAMA KHAN KHOKHAR ENTITLED Differential effects of mutant TAp63γ on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science Madhavi P. Kadakia, Ph.D. Thesis Director Daniel Organisciak , Ph.D. Department Chair Committee on Final Examination Madhavi P. Kadakia, Ph.D. Steven J. Berberich, Ph.D. Michael Leffak, Ph.D. Joseph F. Thomas, Jr., Ph.D. Dean, School of Graduate Studies 3 Abstract Khokhar, Shama K. M.S., Department of Biochemistry and Molecular Biology, Wright State University, 2007 Differential effect of TAp63γ mutants on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression. p63, a member of the p53 gene family, known to play a role in development, has more recently also been implicated in cancer progression. Mice lacking p63 exhibit severe developmental defects such as limb truncations, abnormal skin, and absence of hair follicles, teeth, and mammary glands. Germline missense mutations of p63 have been shown to be responsible for several human developmental syndromes including SHFM, EEC and ADULT syndromes and are associated with anomalies in the development of organs of epithelial origin. -
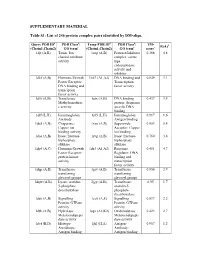
List of 246 Protein Complex Pairs Identified by DM-Align
SUPPLEMENTARY MATERIAL Table S1: List of 246 protein complex pairs identified by DM-align. Query PDB IDa PDB Classb: Temp PDB IDd PDB Classb: TM- R(Å)f (Chain1,Chain2) GO termc (Chain1,Chain2) GO termc scoree 1djt (A,B) Toxin: Ion 1aap (A,B) Protease/Inhibitor 0.368 4.8 channel inhibitor complex: serine activity type endopeptidase activity and inhibitor 1dkf (A,B) Hormone/Growth 1xb7 (A1,A2) DNA binding and 0.849 3.1 Factor Receptor: Transcription DNA binding and factor activity transciption factor activity 1dl5 (A,B) Transferase: 1utx (A,B) DNA binding 0.437 3.9 Methyltransferas protein: Sequence e activity specific DNA binding 1dlf (L,H) Immunoglobin: 1j05 (L,H) Immunoglobin: 0.917 1.6 Antibody Antigen binding 1do5 (A,B) Chaperone: 1xso (A,B) Superoxide 0.953 0.9 Copper ion Acceptor: Copper binding activity ion binding 1dos (A,B) lyase: fructose- 1rvg (A,B) lyase: fructose- 0.760 3.6 biphosphate biphosphate aldolase aldolase 1dp4 (A,C) Hormone/Growth 1dz3 (A1,A2) Response 0.481 4.7 Factor Receptor: Regulator: DNA protein kinase binding and activity transcription factor activity 1dqp (A,B) Transferase: 1grv (A,B) Transferase: 0.856 2.9 transferring transferring glycosyl groups glycosyl groups 1dqw (A,B) Lyase: orotidin- 2jgy (A,B) Transferase: 0.95 1.7 5-phosphate orotidin-5- decarboxylase phosphate decarboxylase 1ds6 (A,B) Signalling 1cc0 (A,E) Signalling 0.897 2.2 Protein: GTPase Protein: GTPase activity activity 1dth (A,B) Hydrolase: 1sqv (A2,K2) Oxidoredutase: 0.423 2.7 Metaloendopepti Metaloendopepti dase activity dase activity 1dvf -

Reduction of Pectinesterase Activity in a Commercial Enzyme Preparation
Journal of the Science of Food and Agriculture J Sci Food Agric 85:1613–1621 (2005) DOI: 10.1002/jsfa.2154 Reduction of pectinesterase activity in a commercial enzyme preparation by pulsed electric fields: comparison of inactivation kinetic models Joaquın´ Giner, Pascal Grouberman, Vicente Gimeno and Olga Martın´ ∗ Department of Food Technology, University of Lleida, CeRTA-UTPV, ETSEA, Avda Alcalde Rovira Roure 191, 25198-Lleida, Spain Abstract: The inactivation of pectinesterase (PE) in a commercial enzyme preparation (CEP) under high intensity pulsed electric fields (HIPEF) was studied. After desalting and water dilution of the raw CEP, samples were exposed to exponentially decay waveform pulses for up to 463 µs at electric field intensities ranging from 19 to 38 kV cm−1. Pulses were applied in monopolar mode. Experimental data were fitted to a first-order kinetic model as well as to models based on Fermi, Hulsheger¨ or Weibull equations to describe PE inactivation kinetics. Characteristic parameters for each model were calculated. Relationships between some of the parameters and process variables were obtained. The Weibull model yielded the best accuracy factor. The relationship between residual PE and input of electrical energy density was found to be that of exponential decay. 2005 Society of Chemical Industry Keywords: pulsed electric fields; kinetics; pectinesterase; model; inactivation INTRODUCTION It has become customary to use CEPs in fruit and Pectinesterase (PE; EC 3.1.1.11) is a pectic enzyme vegetable juice technology. Depending -

W W W .Bio Visio N .Co M New Products Added in 2020
New products added in 2020 Please find below a list of all the products added to our portfolio in the year 2020. Assay Kits Product Name Cat. No. Size Product Name Cat. No. Size N-Acetylcysteine Assay Kit (F) K2044 100 assays Human GAPDH Activity Assay Kit II K2047 100 assays Adeno-Associated Virus qPCR Quantification Kit K1473 100 Rxns Human GAPDH Inhibitor Screening Kit (C) K2043 100 assays 20 Preps, Adenovirus Purification Kit K1459 Hydroxyurea Colorimetric Assay Kit K2046 100 assays 100 Preps Iodide Colorimetric Assay Kit K2037 100 assays Aldehyde Dehydrogenase 2 Inhibitor Screening Kit (F) K2011 100 assays Laccase Activity Assay Kit (C) K2038 100 assays Aldehyde Dehydrogenase 3A1 Inhibitor Screening Kit (F) K2060 100 assays 20 Preps, Lentivirus and Retrovirus Purification Kit K1458 Alkaline Phosphatase Staining Kit K2035 50 assays 100 Preps Alpha-Mannosidase Activity Assay Kit (F) K2041 100 assays Instant Lentivirus Detection Card K1470 10 tests, 20 tests Beta-Mannosidase Activity Assay Kit (F) K2045 100 assays Lentivirus qPCR Quantification Kit K1471 100 Rxns 50 Preps, Buccal Swab DNA Purification Kit K1466 Maleimide Activated KLH-Peptide Conjugation Kit K2039 5 columns 250 Preps Methionine Adenosyltransferase Activity Assay Kit (C) K2033 100 assays CD38 Activity Assay Kit (F) K2042 100 assays miRNA Extraction Kit K1456 50 Preps EZCell™ CFDA SE Cell Tracer Kit K2057 200 assays MMP-13 Inhibitor Screening Kit (F) K2067 100 assays Choline Oxidase Activity Assay Kit (F) K2052 100 assays Mycoplasma PCR Detection Kit K1476 100 Rxns Coronavirus -

Using MALDI-TOF MS Analysis for Rapid Discrimination of MRSA MLST
Using MALDI-TOF MS analysis for rapid discrimination of MRSA MLST types Jang-Jih Lu1,2,Tsui-Ping Liu1, Shih-Cheng Chang1,2 1Department of Laboratory Medicine, Chang Gung Memorial Hospital, Linkou, Taoyuan, Taiwan 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan Email: [email protected] 73 0:F8 MS Ra w 71 0:F9 MS Ra w 5000 6422.794 6000 Intens.[a.u.] Intens.[a.u.] Introduction 4000 Results ST5 ST5 4000 3000 2978.985 2000 6551.531 Staphylococcus aureus is one of the common causes of 3027.802 3054.785 2964.900 3175.943 2000 Table 1: MRSA and MSSA associated peaks by MALDI TOF 1000 community-associated infections and health care related 234 0:F1 MS Ra w 234 0:F1 MS Ra w 3005.919 MRSA(129 isolates) MSSA(77 isolates) 5000 6000 Intens.[a.u.] Intens.[a.u.] pathogen. It can usually cause wound infection, ST45 4000 ST45 6549.206 4000 3000 MS peaks (m/z) no. of isolate % of isolate no. of isolate % of isolate 6420.525 osteomyelitis, and even serious invasive infections, such as 2000 6610.628 2000 2978.373 3053.663 3174.805 2415 67 51.9 2 2.6 1000 sepsis and pneumonia. Epidemiology studies have shown 86 0:E4 MS Ra w 70 0:E5 MS Ra w 3058.565 5000 2431 65 50.4 0 0 3074.390 6000 Intens.[a.u.] Intens.[a.u.] that there are close relationship between the results of ST59 4000 6550.723 ST59 4000 3000 2879 51 39.5 2 2.6 6422.151 genotyping from multilocus sequence typing (MLST) and 2000 2000 characteristics of MRSA strains, including hospital- 6593 77 59.7 7 9.1 1000 200 0:E8 MS Ra w 38 0:E11 MS Ra w Appear of any above 103 79.8 11 14.3 2978.890 5000 6000 Intens.[a.u.] Intens.[a.u.] 6421.658 acquired, community-associated strain and even the 2965.568 ST239 4000 ST239 peaks 4000 3000 none of above peaks 6590.270 26 20.1 66 85.7 2937.113 3177.333 2916.384 antibiogram of the strain. -

HOXB6 Homeo Box B6 HOXB5 Homeo Box B5 WNT5A Wingless-Type
5 6 6 5 . 4 2 1 1 1 2 4 6 4 3 2 9 9 7 0 5 7 5 8 6 4 0 8 2 3 1 8 3 7 1 0 0 4 0 2 5 0 8 7 5 4 1 1 0 3 6 0 4 8 3 7 4 7 6 9 6 7 1 5 0 8 1 4 1 1 7 1 0 0 4 2 0 8 1 1 1 2 5 3 5 0 7 2 6 9 1 2 1 8 3 5 2 9 8 0 6 0 9 5 1 9 9 2 1 1 6 0 2 3 0 3 6 9 1 6 5 5 7 1 1 2 1 1 7 5 4 6 6 4 1 1 2 8 4 7 1 6 2 7 7 5 4 3 2 4 3 6 9 4 1 7 1 3 4 1 2 1 3 1 1 4 7 3 1 1 1 1 5 3 2 6 1 5 1 3 5 4 5 2 3 1 1 6 1 7 3 2 5 4 3 1 6 1 5 3 1 7 6 5 1 1 1 4 6 1 6 2 7 2 1 2 e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S HOXB6 homeo box B6 HOXB5 homeo box B5 WNT5A wingless-type MMTV integration site family, member 5A WNT5A wingless-type MMTV integration site family, member 5A FKBP11 FK506 binding protein 11, 19 kDa EPOR erythropoietin receptor SLC5A6 solute carrier family 5 sodium-dependent vitamin transporter, member 6 SLC5A6 solute carrier family 5 sodium-dependent vitamin transporter, member 6 RAD52 RAD52 homolog S. -

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection.