Updating the Sequence-Based Classification of Glycosyl Hydrolases
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Antifungal Chitosanase from Bacillus Subtilis SH21
molecules Article An Antifungal Chitosanase from Bacillus subtilis SH21 Yuanxiang Pang 1, Jianjun Yang 1, Xinyue Chen 1, Yu Jia 1, Tong Li 1, Junhua Jin 1, Hui Liu 1, Linshu Jiang 1, Yanling Hao 2, Hongxing Zhang 1,* and Yuanhong Xie 1,* 1 Key Laboratory of Agricultural Product Detection and Control of Spoilage Organisms and Pesticides, Beijing Laboratory for Food Quality and Safety, Beijing Engineering Laboratory of Probiotics Key Technology Development, Beijing Engineering Technology Research Center of Food Safety Immune Rapid Detection, Food Science and Engineering College, Beijing University of Agriculture, Beijing 102206, China; [email protected] (Y.P.); [email protected] (J.Y.); [email protected] (X.C.); [email protected] (Y.J.); [email protected] (T.L.); [email protected] (J.J.); [email protected] (H.L.); [email protected] (L.J.) 2 Key Laboratory of Functional Dairy Science of Beijing and Chinese Ministry of Education, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing 100083, China; [email protected] * Correspondence: [email protected] (H.Z.); [email protected] (Y.X.) Abstract: Bacillus subtilis SH21 was observed to produce an antifungal protein that inhibited the growth of F. solani. To purify this protein, ammonium sulfate precipitation, gel filtration chromatogra- phy, and ion-exchange chromatography were used. The purity of the purified product was 91.33% ac- cording to high-performance liquid chromatography results. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis revealed that the molecular weight of the protein is 30.72 kDa. The results of the LC–MS/MS analy- sis and a subsequent sequence-database search indicated that this protein was a chitosanase, and thus, we named it chitosanase SH21. -

THE ALPHA-GALACTOSIDASE SUPERFAMILY: SEQUENCE BASED CLASSIFICATION of ALPHA-GALACTOSIDASES and RELATED GLYCOSIDASES Naumoff D.G
COMPUTATIONAL STRUCTURAL AND FUNCTIONAL PROTEOMICS THE ALPHA-GALACTOSIDASE SUPERFAMILY: SEQUENCE BASED CLASSIFICATION OF ALPHA-GALACTOSIDASES AND RELATED GLYCOSIDASES Naumoff D.G. State Institute for Genetics and Selection of Industrial Microorganisms, Moscow, Russia, e-mail: [email protected] Keywords: α-galactosidase, melibiase, glycoside hydrolase, GH-D clan, GH31 family, GHX family, COG1649, enzyme classification, protein family, protein phylogeny Summary Motivation: About 1 % of genes in genomes code enzymes with glycosidase activities. On the basis of sequence similarity all known glycosidases have been classified into 90 families. In many cases proteins of different families have common evolution origin. It makes necessary to combine the corresponding families into a superfamily. Results: Using of the PSI-BLAST program we found significant sequence similarity of several glycosidase families, two of which includes enzymes with the α galactosidase activity. Sequence homology, common catalytic mechanism, folding similarities, and composition of the active center allowed us to group three of these families – GH27, GH31, and GH36 – into the α-galactosidase superfamily. Phylogenetic analysis of this superfamily revealed polyphyletic origin of GH36 family, which could be divided into four families. Glycosidases of the α-galactosidase superfamily have a distant relationship with proteins belonging to families GH13, GH70, and GH77 of glycosidases, as well as with two families of predicted glycosidases. Introduction Glycoside hydrolases or glycosidases (EC 3.2.1.-) are a widespread group of enzymes, hydrolyzing the glycosidic bonds between two carbohydrates or between a carbohydrate and an aglycone moiety. A large multiplicity of these enzymes is a consequence of the extensive variety of their natural substrates: di-, oligo-, and polysaccharides. -

Depolymerization of Chitosan by Enzymes from the Digestive Tract of Sea Cucumber Stichopus Japonicus
African Journal of Biotechnology Vol. 11(2), pp. 423-428, 5 January, 2012 Available online at http://www.academicjournals.org/AJB DOI: 10.5897/AJB11.2803 ISSN 1684–5315 © 2012 Academic Journals Full Length Research Paper Depolymerization of chitosan by enzymes from the digestive tract of sea cucumber Stichopus japonicus Dong-Rui Yao, Ming-Qian Zhou, Sheng-Jun Wu and Sai-Kun Pan* School of Marine Science and Technology, Huaihai Institute of Technology, 59 Cangwu Road, Xinpu, 222005, China. Accepted 14 November, 2011 A complex of enzymes was isolated in a preparation derived from the digestive tract of sea cucumber, Stichopus japonicus . Hydrolysis of chitosan using this enzyme preparation decreased its molecular weight (Mw), increased its water solubility and produced water-soluble chitosan (WSC). The conditions for hydrolysis were optimized to pH 6.0, temperature 45°C, 16 mg enzyme preparation (22.08 U of chitosanase activity) in a reaction solution (500 ml) containing 5 g chitosan and total reaction time of 3 h. The Mw of hydrolyzed chitosan was 1260 Da, and the WSC content in the resulting product and the yield were 96.7 and 95.4% (w/w), respectively. The structure of WSC was characterized using Fourier transform infrared (FTIR) spectroscopy. Key words: Water-soluble chitosan, complex enzyme preparation, sea cucumber Stichopus japonicus , hydrolysis. INTRODUCTION Water-soluble chitosan (WSC) has many advantages 2007). when compared with ordinary chitosan; these include Furthermore, a number of enzymes, such as protease, antifungal, antibacterial and antitumor activities (Kang et lipase, esterase, glycosidase (amylase, cellulose, dis- al., 2007). WSC can be synthesized by either chemical or accharidases, invertase and chitinase) and phosphatase enzymatic hydrolysis. -

New Insights Into the Action of Bacterial Chondroitinase AC I And
Carbohydrate Polymers 158 (2017) 85–92 Contents lists available at ScienceDirect Carbohydrate Polymers journal homepage: www.elsevier.com/locate/carbpol New insights into the action of bacterial chondroitinase AC I and hyaluronidase on hyaluronic acid a a a,∗ a b a,∗ Lei Tao , Fei Song , Naiyu Xu , Duxin Li , Robert J. Linhardt , Zhenqing Zhang a Jiangsu Key Laboratory of Translational Research and Therapy for Neuro-Psycho-Diseases and College of Pharmaceutical Sciences, Soochow University, Suzhou, Jiangsu 215021, China b Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, 110 8th St., Troy, NY, 12180, USA a r t i c l e i n f o a b s t r a c t Article history: Hyaluronic acid (HA), a glycosaminoglycan, is a linear polysaccharide with negative charge, Received 19 October 2016 composed of a repeating disaccharide unit [→4)--d-glucopyranosyluronic acid (1 → 3)--d- N-acetyl-d- Received in revised form 2 December 2016 glucoaminopyranose (1 → ]n ([→4) GlcA (1 → 3) GlcNAc 1 → ]n). It is widely used in different applications Accepted 5 December 2016 based on its physicochemical properties associated with its molecular weight. Enzymatic digestion by Available online 6 December 2016 polysaccharides lyases is one of the most important ways to decrease the molecular weight of HA. Thus, it is important to understand the action patterns of lyases acting on HA. In this study, the action pat- Keywords: terns of two common lyases, Flavobacterial chondroitinase AC I and Streptomyces hyaluronidase, were Hyaluronic acid investigated by analyzing HA oligosaccharide digestion products. HA oligosaccharides having an odd- Chondroitinase AC Hyaluronidase number of saccharide residues were observed in the products of both lyases, but their distributions were  Odd-numbered oligosaccharides quite different. -

Improve Thermostability of Bacillus Sp. TS Chitosanase Through Structure‑Based Alignment Zhanping Zhou1 & Xiao Wang2*
www.nature.com/scientificreports OPEN Improve thermostability of Bacillus sp. TS chitosanase through structure‑based alignment Zhanping Zhou1 & Xiao Wang2* Chitosanases can catalyze the release of chitooligosaccharides which have a number of medical applications. Therefore, Chitosanases are good candidates for large‑scale enzymatic synthesis due to their favorable thermostability properties and high catalytic efciency. To further improve the thermostability of a chitosanase from Bacillus sp. TS, which has a half‑life of 5.32 min, we mutated specifc serine residues that we identifed as potentially relevant through structure comparison with thermophilic CelA from Clostridium thermocellum. Out of a total of 15 mutants, three, namely S265G, S276A, and S347G, show higher thermostability. Their half‑lives at 60 °C were calculated as 34.57 min, 36.79 min and 7.2 min. The Km values of S265G, S276A and S347G mutants show substrate binding ability comparable to that of the wild‑type enzyme, while the S265G mutant displays a signifcant decrease of enzymatic activities. Additionally, we studied the synergistic efects of combined mutations, observing that all double mutants and the triple mutant are more stable than the wild‑type enzyme and single mutants. Finally, we investigated the mechanisms which might give a reasonable explanation for the improved thermostability via comparative analysis of the resulting 3D structures. Chitosanase (EC 3.2.1.132) is a kind of enzyme that can degrade chitosan and produce chitooligosaccharides 1, which are mainly composed of D-glucosamine and occasionally incorporation of N-acetylglucosamine. Te chitooligosaccharides exhibit a wide range of interesting biological activities. A number of potential applications of chitooligosaccharides have been suggested in medical contexts, including their use as inhibitors of tumor growth and metastasis, anti-infammation therapeutics against asthma, facilitators of bone-tissue formation, wound healing accelerators and antibacterial, antifungal, and anti-malaria agents2–4. -

Discovery, Molecular Mechanisms, and Industrial Applications of Cold-Active Enzymes
REVIEW published: 09 September 2016 doi: 10.3389/fmicb.2016.01408 Discovery, Molecular Mechanisms, and Industrial Applications of Cold-Active Enzymes Margarita Santiago 1, César A. Ramírez-Sarmiento 2, Ricardo A. Zamora 3 and Loreto P. Parra 2, 4* 1 Department of Chemical Engineering and Biotechnology, Centre for Biochemical Engineering and Biotechnology, Universidad de Chile, Santiago, Chile, 2 Schools of Engineering, Medicine and Biological Sciences, Institute for Biological and Medical Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 3 Departamento de Biología, Facultad de Ciencias, Universidad de Chile, Santiago, Chile, 4 Department of Chemical and Bioprocesses Engineering, School of Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile Cold-active enzymes constitute an attractive resource for biotechnological applications. Their high catalytic activity at temperatures below 25◦C makes them excellent biocatalysts that eliminate the need of heating processes hampering the quality, sustainability, and cost-effectiveness of industrial production. Here we provide a review of the isolation and characterization of novel cold-active enzymes from microorganisms Edited by: inhabiting different environments, including a revision of the latest techniques that have Robert Kourist, been used for accomplishing these paramount tasks. We address the progress made Ruhr University Bochum, Germany in the overexpression and purification of cold-adapted enzymes, the evolutionary and Reviewed by: molecular basis of their high activity at low temperatures and the experimental and Kerstin Steiner, Austrian Centre of Industrial computational techniques used for their identification, along with protein engineering Biotechnology, Austria endeavors based on these observations to improve some of the properties of Sandy Schmidt, Delft University of Technology, cold-adapted enzymes to better suit specific applications. -

Immobilization of -Galactosidases on the Lactobacillus Cell Surface
catalysts Article Immobilization of β-Galactosidases on the Lactobacillus Cell Surface Using the Peptidoglycan-Binding Motif LysM Mai-Lan Pham 1 , Anh-Minh Tran 1,2, Suwapat Kittibunchakul 1, Tien-Thanh Nguyen 3, Geir Mathiesen 4 and Thu-Ha Nguyen 1,* 1 Food Biotechnology Laboratory, Department of Food Science and Technology, BOKU-University of Natural Resources and Life Sciences, A-1190 Vienna, Austria; [email protected] (M.-L.P.); [email protected] (A.-M.T.); [email protected] (S.K.) 2 Department of Biology, Faculty of Fundamental Sciences, Ho Chi Minh City University of Medicine and Pharmacy, 217 Hong Bang, Ho Chi Minh City, Vietnam 3 School of Biotechnology and Food Technology, Hanoi University of Science and Technology, 1 Dai Co Viet, Hanoi, Vietnam; [email protected] 4 Faculty of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences (NMBU), N-1432 Ås, Norway; [email protected] * Correspondence: [email protected]; Tel.: +43-1-47654-75215; Fax: +43-1-47654-75039 Received: 25 April 2019; Accepted: 7 May 2019; Published: 12 May 2019 Abstract: Lysin motif (LysM) domains are found in many bacterial peptidoglycan hydrolases. They can bind non-covalently to peptidoglycan and have been employed to display heterologous proteins on the bacterial cell surface. In this study, we aimed to use a single LysM domain derived from a putative extracellular transglycosylase Lp_3014 of Lactobacillus plantarum WCFS1 to display two different lactobacillal β-galactosidases, the heterodimeric LacLM-type from Lactobacillus reuteri and the homodimeric LacZ-type from Lactobacillus delbrueckii subsp. bulgaricus, on the cell surface of different Lactobacillus spp. -

Supplemental Table S1: Comparison of the Deleted Genes in the Genome-Reduced Strains
Supplemental Table S1: Comparison of the deleted genes in the genome-reduced strains Legend 1 Locus tag according to the reference genome sequence of B. subtilis 168 (NC_000964) Genes highlighted in blue have been deleted from the respective strains Genes highlighted in green have been inserted into the indicated strain, they are present in all following strains Regions highlighted in red could not be deleted as a unit Regions highlighted in orange were not deleted in the genome-reduced strains since their deletion resulted in severe growth defects Gene BSU_number 1 Function ∆6 IIG-Bs27-47-24 PG10 PS38 dnaA BSU00010 replication initiation protein dnaN BSU00020 DNA polymerase III (beta subunit), beta clamp yaaA BSU00030 unknown recF BSU00040 repair, recombination remB BSU00050 involved in the activation of biofilm matrix biosynthetic operons gyrB BSU00060 DNA-Gyrase (subunit B) gyrA BSU00070 DNA-Gyrase (subunit A) rrnO-16S- trnO-Ala- trnO-Ile- rrnO-23S- rrnO-5S yaaC BSU00080 unknown guaB BSU00090 IMP dehydrogenase dacA BSU00100 penicillin-binding protein 5*, D-alanyl-D-alanine carboxypeptidase pdxS BSU00110 pyridoxal-5'-phosphate synthase (synthase domain) pdxT BSU00120 pyridoxal-5'-phosphate synthase (glutaminase domain) serS BSU00130 seryl-tRNA-synthetase trnSL-Ser1 dck BSU00140 deoxyadenosin/deoxycytidine kinase dgk BSU00150 deoxyguanosine kinase yaaH BSU00160 general stress protein, survival of ethanol stress, SafA-dependent spore coat yaaI BSU00170 general stress protein, similar to isochorismatase yaaJ BSU00180 tRNA specific adenosine -

Gene Cloning, Functional Expression and Characterization of a Novel GH46 Chitosanase from Streptomyces Avermitilis (Sacsn46a)
Gene cloning, functional expression and characterization of a novel GH46 chitosanase from Streptomyces avermitilis (SaCsn46A) Jing Guo ( [email protected] ) Changzhou University https://orcid.org/0000-0002-7622-6810 Yi Wang Changzhou University Wenjun Gao Changzhou University Xinrou Wang Changzhou University Xin Gao Changzhou University Zaiwei Man Changzhou University Zhiqiang Cai Changzhou University Qing Qing Changzhou University Research Article Keywords: Streptomyces avermitilis, Chitosanase, Glucosamine, Chitooligosaccharides, Glycoside hydrolase (GH) family 46, Enzyme properties Posted Date: August 11th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-785979/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/17 Abstract A novel glycoside hydrolase (GH) family 46 chitosanase (SaCsn46A) from Streptomyces avermitilis was cloned and functionally expressed in Escherichia coli Rosetta (DE3) strains. SaCsn46A consists of 271 amino acids, which includes a 34-amino acids signal peptide. The protein sequence of SaCsn46A shows maximum identity (83.5%) to chitosanase from Streptomyces sp. SirexAA-E. Then the mature enzyme was puried to homogeneity through Ni-chelating anity chromatography with a recovery yield of 78% and the molecular mass of puried enzyme was estimated to be 29 kDa by SDS-PAGE. The recombinant enzyme possessed a temperature optimum of 45 oC and a pH optimum of 6.2, and it was stable at pH o − 1 ranging from 4.0 to 9.0 and below 30 C. The Km and Vmax values of this enzyme were 1.32 mg∙mL , 526.32 µM∙mg− 1∙min− 1, respectively (chitosan as substrate). The enzyme activity can be enhanced by Mg2+ and especially Mn2+, which could enhance the activity about 3.62-fold at a 3 mM concentration. -
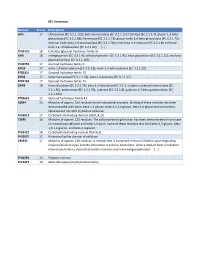
Beta-Mannosidase (EC 3.2.1.25)
M1 Consensus Domain # runs Description GH5 17 chitosanase (EC 3.2.1.132); beta-mannosidase (EC 3.2.1.25); Cellulase (EC 3.2.1.4); glucan 1,3-beta- glucosidase (EC 3.2.1.58); licheninase (EC 3.2.1.73); glucan endo-1,6-beta-glucosidase (EC 3.2.1.75); mannan endo-beta-1,4-mannosidase (EC 3.2.1.78); endo-beta-1,4-xylanase (EC 3.2.1.8); cellulose beta-1,4-cellobiosidase (EC 3.2.1.91); [...] PF00150 18 Cellulase (glycosyl hydrolase family 5) GH9 18 endoglucanase (EC 3.2.1.4); cellobiohydrolase (EC 3.2.1.91); beta-glucosidase (EC 3.2.1.21); exo-beta- glucosaminidase (EC 3.2.1.165) PF00759 17 Glycosyl hydrolase family 9 GH10 17 endo-1,4-beta-xylanase (EC 3.2.1.8); endo-1,3-beta-xylanase (EC 3.2.1.32) PF00331 17 Glycosyl hydrolase family 10 GH26 17 beta-mannanase (EC 3.2.1.78); beta-1,3-xylanase (EC 3.2.1.32) PF02156 17 Glycosyl hydrolase family 26 GH43 18 beta-xylosidase (EC 3.2.1.37); beta-1,3-xylosidase (EC 3.2.1.-); alpha-L-arabinofuranosidase (EC 3.2.1.55); arabinanase (EC 3.2.1.99); xylanase (EC 3.2.1.8); galactan 1,3-beta-galactosidase (EC 3.2.1.145) PF04616 17 Glycosyl hydrolases family 43 CBM4 16 Modules of approx. 150 residues found in bacterial enzymes. Binding of these modules has been demonstrated with xylan, beta-1,3-glucan, beta-1,3-1,4-glucan, beta-1,6-glucan and amorphous cellulose but not with crystalline cellulose. -

Amylases and Related Glycoside Hydrolases with Transglycosylation
Amylase 2018; 2: 17–29 Review Article Mary Casa-Villegas, Julia Marín-Navarro, Julio Polaina* Amylases and related glycoside hydrolases with transglycosylation activity used for the production of isomaltooligosaccharides https://doi.org/10.1515/amylase-2018-0003 Abbreviations: AIG, α-glucosidase from Acremonium Received March 26, 2018; accepted May 20, 2018 implicatum; ANG, α-glucosidase from Aspergillus niger; Abstract: Isomaltooligosaccharides (IMOS) are sugars AOG, α-glucosidase from Aspergillus oryzae; BcCITase, with health promoting properties that make them relevant CITase from Bacillus circulans T-3040; CBM, carbohydrate- for the pharmaceutical and food industries. IMOS have binding module; CI, cyclic isomaltooligosaccharide; ample chemical diversity achieved by different α-glucosidic CITase, cycloisomaltooligosaccharide glucanotransferase; linkages and polymerization degrees, forming linear, CMM, cyclic maltosylmaltose; CTS, cyclic tetrasaccharide; branched and cyclic structures. Enzymatic synthesis of DP, degree of polymerization; FOS, fructooligosaccharides; these compounds can be carried out by glycoside hydrolases GH, glycoside hydrolases; GOS, galactooligosaccharides; (GHs) with transglycosylating activity. Different substrates IMOS, isomaltooligosaccharides; OPMA-N, maltogenic are used for the synthesis: combinations of disaccharides amylase from Bacillus sp.; PsCITase, CITase from and monosaccharides, or polymeric carbohydrates such Paenibacillus sp. 598K; SI, sucrose isomerase; SmuA, as starch or dextran, which are converted -

Immobilization of Dextranase on Nano-Hydroxyapatite As a Recyclable Catalyst
materials Article Immobilization of Dextranase on Nano-Hydroxyapatite as a Recyclable Catalyst Yanshuai Ding 1,2 , Hao Zhang 1,2 , Xuelian Wang 1,2, Hangtian Zu 1,2 , Cang Wang 1,2, Dongxue Dong 1,2, Mingsheng Lyu 1,2,3 and Shujun Wang 1,2,3,* 1 Jiangsu Key Laboratory of Marine Bioresources and Environment, Jiangsu Ocean University, Lianyungang 222005, China; [email protected] (Y.D.); [email protected] (H.Z.); [email protected] (X.W.); [email protected] (H.Z.); [email protected] (C.W.); [email protected] (D.D.); [email protected] (M.L.) 2 Co-Innovation Center of Jiangsu Marine Bio-industry Technology, Jiangsu Ocean University, Lianyungang 222005, China 3 Collaborative Innovation Center of Modern Biological Manufacturing, Anhui University, Hefei 230039, China * Correspondence: [email protected] Abstract: The immobilization technology provides a potential pathway for enzyme recycling. Here, we evaluated the potential of using dextranase immobilized onto hydroxyapatite nanoparticles as a promising inorganic material. The optimal immobilization temperature, reaction time, and pH were determined to be 25 ◦C, 120 min, and pH 5, respectively. Dextranase could be loaded at 359.7 U/g. The immobilized dextranase was characterized by field emission gun-scanning electron microscope (FEG-SEM), X-ray diffraction (XRD), and Fourier-transformed infrared spectroscopy (FT-IR). The hydrolysis capacity of the immobilized enzyme was maintained at 71% at the 30th time of use. According to the constant temperature acceleration experiment, it was estimated that the immobilized dextranase could be stored for 99 days at 20 ◦C, indicating that the immobilized enzyme had good storage properties.