Secretory Production of a Beta-Mannanase and A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Antifungal Chitosanase from Bacillus Subtilis SH21
molecules Article An Antifungal Chitosanase from Bacillus subtilis SH21 Yuanxiang Pang 1, Jianjun Yang 1, Xinyue Chen 1, Yu Jia 1, Tong Li 1, Junhua Jin 1, Hui Liu 1, Linshu Jiang 1, Yanling Hao 2, Hongxing Zhang 1,* and Yuanhong Xie 1,* 1 Key Laboratory of Agricultural Product Detection and Control of Spoilage Organisms and Pesticides, Beijing Laboratory for Food Quality and Safety, Beijing Engineering Laboratory of Probiotics Key Technology Development, Beijing Engineering Technology Research Center of Food Safety Immune Rapid Detection, Food Science and Engineering College, Beijing University of Agriculture, Beijing 102206, China; [email protected] (Y.P.); [email protected] (J.Y.); [email protected] (X.C.); [email protected] (Y.J.); [email protected] (T.L.); [email protected] (J.J.); [email protected] (H.L.); [email protected] (L.J.) 2 Key Laboratory of Functional Dairy Science of Beijing and Chinese Ministry of Education, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing 100083, China; [email protected] * Correspondence: [email protected] (H.Z.); [email protected] (Y.X.) Abstract: Bacillus subtilis SH21 was observed to produce an antifungal protein that inhibited the growth of F. solani. To purify this protein, ammonium sulfate precipitation, gel filtration chromatogra- phy, and ion-exchange chromatography were used. The purity of the purified product was 91.33% ac- cording to high-performance liquid chromatography results. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis revealed that the molecular weight of the protein is 30.72 kDa. The results of the LC–MS/MS analy- sis and a subsequent sequence-database search indicated that this protein was a chitosanase, and thus, we named it chitosanase SH21. -

Updating the Sequence-Based Classification of Glycosyl Hydrolases
Article Updating the sequence-based classification of glycosyl hydrolases HENRISSAT, Bernard, BAIROCH, Amos Marc Reference HENRISSAT, Bernard, BAIROCH, Amos Marc. Updating the sequence-based classification of glycosyl hydrolases. Biochemical Journal, 1996, vol. 316 ( Pt 2), p. 695-6 PMID : 8687420 DOI : 10.1042/bj3160695 Available at: http://archive-ouverte.unige.ch/unige:36909 Disclaimer: layout of this document may differ from the published version. 1 / 1 Biochem. J. (1996) 316, 695–696 (Printed in Great Britain) 695 BIOCHEMICAL JOURNAL Updating the sequence-based classification of available. When the number of glycosyl hydrolase sequences reached C 480, ten additional families (designated 36–45) could glycosyl hydrolases be defined and were added to the classification [2]. There are at present over 950 sequences of glycosyl hydrolases in the data- A classification of glycosyl hydrolases based on amino-acid- banks (EMBL}GenBank and SWISS-PROT). Their analysis sequence similarities was proposed in this Journal a few years shows that the vast majority of the C 470 additional sequences ago [1]. This classification originated from the analysis of C 300 that have become available since the last update could be classified sequences and their grouping into 35 families designated 1–35. in the existing families. However, several sequences not fitting Because such a classification is necessarily sensitive to the sample, the existing families allow the definition of new families (desig- it was anticipated that it was incomplete and that new families nated 46–57) (Table 1). When the several present genome would be determined when additional sequences would become sequencing projects have reached completion, the number of Table 1 New families in the classification of glycosyl hydrolases Family Enzyme Organism SWISS-PROT EMBL/GenBank 46 Chitosanase Bacillus circulans MH-K1 P33673 D10624 46 Chitosanase Streptomyces sp. -

Depolymerization of Chitosan by Enzymes from the Digestive Tract of Sea Cucumber Stichopus Japonicus
African Journal of Biotechnology Vol. 11(2), pp. 423-428, 5 January, 2012 Available online at http://www.academicjournals.org/AJB DOI: 10.5897/AJB11.2803 ISSN 1684–5315 © 2012 Academic Journals Full Length Research Paper Depolymerization of chitosan by enzymes from the digestive tract of sea cucumber Stichopus japonicus Dong-Rui Yao, Ming-Qian Zhou, Sheng-Jun Wu and Sai-Kun Pan* School of Marine Science and Technology, Huaihai Institute of Technology, 59 Cangwu Road, Xinpu, 222005, China. Accepted 14 November, 2011 A complex of enzymes was isolated in a preparation derived from the digestive tract of sea cucumber, Stichopus japonicus . Hydrolysis of chitosan using this enzyme preparation decreased its molecular weight (Mw), increased its water solubility and produced water-soluble chitosan (WSC). The conditions for hydrolysis were optimized to pH 6.0, temperature 45°C, 16 mg enzyme preparation (22.08 U of chitosanase activity) in a reaction solution (500 ml) containing 5 g chitosan and total reaction time of 3 h. The Mw of hydrolyzed chitosan was 1260 Da, and the WSC content in the resulting product and the yield were 96.7 and 95.4% (w/w), respectively. The structure of WSC was characterized using Fourier transform infrared (FTIR) spectroscopy. Key words: Water-soluble chitosan, complex enzyme preparation, sea cucumber Stichopus japonicus , hydrolysis. INTRODUCTION Water-soluble chitosan (WSC) has many advantages 2007). when compared with ordinary chitosan; these include Furthermore, a number of enzymes, such as protease, antifungal, antibacterial and antitumor activities (Kang et lipase, esterase, glycosidase (amylase, cellulose, dis- al., 2007). WSC can be synthesized by either chemical or accharidases, invertase and chitinase) and phosphatase enzymatic hydrolysis. -

Improve Thermostability of Bacillus Sp. TS Chitosanase Through Structure‑Based Alignment Zhanping Zhou1 & Xiao Wang2*
www.nature.com/scientificreports OPEN Improve thermostability of Bacillus sp. TS chitosanase through structure‑based alignment Zhanping Zhou1 & Xiao Wang2* Chitosanases can catalyze the release of chitooligosaccharides which have a number of medical applications. Therefore, Chitosanases are good candidates for large‑scale enzymatic synthesis due to their favorable thermostability properties and high catalytic efciency. To further improve the thermostability of a chitosanase from Bacillus sp. TS, which has a half‑life of 5.32 min, we mutated specifc serine residues that we identifed as potentially relevant through structure comparison with thermophilic CelA from Clostridium thermocellum. Out of a total of 15 mutants, three, namely S265G, S276A, and S347G, show higher thermostability. Their half‑lives at 60 °C were calculated as 34.57 min, 36.79 min and 7.2 min. The Km values of S265G, S276A and S347G mutants show substrate binding ability comparable to that of the wild‑type enzyme, while the S265G mutant displays a signifcant decrease of enzymatic activities. Additionally, we studied the synergistic efects of combined mutations, observing that all double mutants and the triple mutant are more stable than the wild‑type enzyme and single mutants. Finally, we investigated the mechanisms which might give a reasonable explanation for the improved thermostability via comparative analysis of the resulting 3D structures. Chitosanase (EC 3.2.1.132) is a kind of enzyme that can degrade chitosan and produce chitooligosaccharides 1, which are mainly composed of D-glucosamine and occasionally incorporation of N-acetylglucosamine. Te chitooligosaccharides exhibit a wide range of interesting biological activities. A number of potential applications of chitooligosaccharides have been suggested in medical contexts, including their use as inhibitors of tumor growth and metastasis, anti-infammation therapeutics against asthma, facilitators of bone-tissue formation, wound healing accelerators and antibacterial, antifungal, and anti-malaria agents2–4. -

Discovery, Molecular Mechanisms, and Industrial Applications of Cold-Active Enzymes
REVIEW published: 09 September 2016 doi: 10.3389/fmicb.2016.01408 Discovery, Molecular Mechanisms, and Industrial Applications of Cold-Active Enzymes Margarita Santiago 1, César A. Ramírez-Sarmiento 2, Ricardo A. Zamora 3 and Loreto P. Parra 2, 4* 1 Department of Chemical Engineering and Biotechnology, Centre for Biochemical Engineering and Biotechnology, Universidad de Chile, Santiago, Chile, 2 Schools of Engineering, Medicine and Biological Sciences, Institute for Biological and Medical Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 3 Departamento de Biología, Facultad de Ciencias, Universidad de Chile, Santiago, Chile, 4 Department of Chemical and Bioprocesses Engineering, School of Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile Cold-active enzymes constitute an attractive resource for biotechnological applications. Their high catalytic activity at temperatures below 25◦C makes them excellent biocatalysts that eliminate the need of heating processes hampering the quality, sustainability, and cost-effectiveness of industrial production. Here we provide a review of the isolation and characterization of novel cold-active enzymes from microorganisms Edited by: inhabiting different environments, including a revision of the latest techniques that have Robert Kourist, been used for accomplishing these paramount tasks. We address the progress made Ruhr University Bochum, Germany in the overexpression and purification of cold-adapted enzymes, the evolutionary and Reviewed by: molecular basis of their high activity at low temperatures and the experimental and Kerstin Steiner, Austrian Centre of Industrial computational techniques used for their identification, along with protein engineering Biotechnology, Austria endeavors based on these observations to improve some of the properties of Sandy Schmidt, Delft University of Technology, cold-adapted enzymes to better suit specific applications. -

Immobilization of -Galactosidases on the Lactobacillus Cell Surface
catalysts Article Immobilization of β-Galactosidases on the Lactobacillus Cell Surface Using the Peptidoglycan-Binding Motif LysM Mai-Lan Pham 1 , Anh-Minh Tran 1,2, Suwapat Kittibunchakul 1, Tien-Thanh Nguyen 3, Geir Mathiesen 4 and Thu-Ha Nguyen 1,* 1 Food Biotechnology Laboratory, Department of Food Science and Technology, BOKU-University of Natural Resources and Life Sciences, A-1190 Vienna, Austria; [email protected] (M.-L.P.); [email protected] (A.-M.T.); [email protected] (S.K.) 2 Department of Biology, Faculty of Fundamental Sciences, Ho Chi Minh City University of Medicine and Pharmacy, 217 Hong Bang, Ho Chi Minh City, Vietnam 3 School of Biotechnology and Food Technology, Hanoi University of Science and Technology, 1 Dai Co Viet, Hanoi, Vietnam; [email protected] 4 Faculty of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences (NMBU), N-1432 Ås, Norway; [email protected] * Correspondence: [email protected]; Tel.: +43-1-47654-75215; Fax: +43-1-47654-75039 Received: 25 April 2019; Accepted: 7 May 2019; Published: 12 May 2019 Abstract: Lysin motif (LysM) domains are found in many bacterial peptidoglycan hydrolases. They can bind non-covalently to peptidoglycan and have been employed to display heterologous proteins on the bacterial cell surface. In this study, we aimed to use a single LysM domain derived from a putative extracellular transglycosylase Lp_3014 of Lactobacillus plantarum WCFS1 to display two different lactobacillal β-galactosidases, the heterodimeric LacLM-type from Lactobacillus reuteri and the homodimeric LacZ-type from Lactobacillus delbrueckii subsp. bulgaricus, on the cell surface of different Lactobacillus spp. -

Supplemental Table S1: Comparison of the Deleted Genes in the Genome-Reduced Strains
Supplemental Table S1: Comparison of the deleted genes in the genome-reduced strains Legend 1 Locus tag according to the reference genome sequence of B. subtilis 168 (NC_000964) Genes highlighted in blue have been deleted from the respective strains Genes highlighted in green have been inserted into the indicated strain, they are present in all following strains Regions highlighted in red could not be deleted as a unit Regions highlighted in orange were not deleted in the genome-reduced strains since their deletion resulted in severe growth defects Gene BSU_number 1 Function ∆6 IIG-Bs27-47-24 PG10 PS38 dnaA BSU00010 replication initiation protein dnaN BSU00020 DNA polymerase III (beta subunit), beta clamp yaaA BSU00030 unknown recF BSU00040 repair, recombination remB BSU00050 involved in the activation of biofilm matrix biosynthetic operons gyrB BSU00060 DNA-Gyrase (subunit B) gyrA BSU00070 DNA-Gyrase (subunit A) rrnO-16S- trnO-Ala- trnO-Ile- rrnO-23S- rrnO-5S yaaC BSU00080 unknown guaB BSU00090 IMP dehydrogenase dacA BSU00100 penicillin-binding protein 5*, D-alanyl-D-alanine carboxypeptidase pdxS BSU00110 pyridoxal-5'-phosphate synthase (synthase domain) pdxT BSU00120 pyridoxal-5'-phosphate synthase (glutaminase domain) serS BSU00130 seryl-tRNA-synthetase trnSL-Ser1 dck BSU00140 deoxyadenosin/deoxycytidine kinase dgk BSU00150 deoxyguanosine kinase yaaH BSU00160 general stress protein, survival of ethanol stress, SafA-dependent spore coat yaaI BSU00170 general stress protein, similar to isochorismatase yaaJ BSU00180 tRNA specific adenosine -

Gene Cloning, Functional Expression and Characterization of a Novel GH46 Chitosanase from Streptomyces Avermitilis (Sacsn46a)
Gene cloning, functional expression and characterization of a novel GH46 chitosanase from Streptomyces avermitilis (SaCsn46A) Jing Guo ( [email protected] ) Changzhou University https://orcid.org/0000-0002-7622-6810 Yi Wang Changzhou University Wenjun Gao Changzhou University Xinrou Wang Changzhou University Xin Gao Changzhou University Zaiwei Man Changzhou University Zhiqiang Cai Changzhou University Qing Qing Changzhou University Research Article Keywords: Streptomyces avermitilis, Chitosanase, Glucosamine, Chitooligosaccharides, Glycoside hydrolase (GH) family 46, Enzyme properties Posted Date: August 11th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-785979/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/17 Abstract A novel glycoside hydrolase (GH) family 46 chitosanase (SaCsn46A) from Streptomyces avermitilis was cloned and functionally expressed in Escherichia coli Rosetta (DE3) strains. SaCsn46A consists of 271 amino acids, which includes a 34-amino acids signal peptide. The protein sequence of SaCsn46A shows maximum identity (83.5%) to chitosanase from Streptomyces sp. SirexAA-E. Then the mature enzyme was puried to homogeneity through Ni-chelating anity chromatography with a recovery yield of 78% and the molecular mass of puried enzyme was estimated to be 29 kDa by SDS-PAGE. The recombinant enzyme possessed a temperature optimum of 45 oC and a pH optimum of 6.2, and it was stable at pH o − 1 ranging from 4.0 to 9.0 and below 30 C. The Km and Vmax values of this enzyme were 1.32 mg∙mL , 526.32 µM∙mg− 1∙min− 1, respectively (chitosan as substrate). The enzyme activity can be enhanced by Mg2+ and especially Mn2+, which could enhance the activity about 3.62-fold at a 3 mM concentration. -
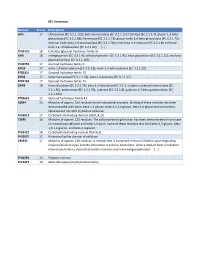
Beta-Mannosidase (EC 3.2.1.25)
M1 Consensus Domain # runs Description GH5 17 chitosanase (EC 3.2.1.132); beta-mannosidase (EC 3.2.1.25); Cellulase (EC 3.2.1.4); glucan 1,3-beta- glucosidase (EC 3.2.1.58); licheninase (EC 3.2.1.73); glucan endo-1,6-beta-glucosidase (EC 3.2.1.75); mannan endo-beta-1,4-mannosidase (EC 3.2.1.78); endo-beta-1,4-xylanase (EC 3.2.1.8); cellulose beta-1,4-cellobiosidase (EC 3.2.1.91); [...] PF00150 18 Cellulase (glycosyl hydrolase family 5) GH9 18 endoglucanase (EC 3.2.1.4); cellobiohydrolase (EC 3.2.1.91); beta-glucosidase (EC 3.2.1.21); exo-beta- glucosaminidase (EC 3.2.1.165) PF00759 17 Glycosyl hydrolase family 9 GH10 17 endo-1,4-beta-xylanase (EC 3.2.1.8); endo-1,3-beta-xylanase (EC 3.2.1.32) PF00331 17 Glycosyl hydrolase family 10 GH26 17 beta-mannanase (EC 3.2.1.78); beta-1,3-xylanase (EC 3.2.1.32) PF02156 17 Glycosyl hydrolase family 26 GH43 18 beta-xylosidase (EC 3.2.1.37); beta-1,3-xylosidase (EC 3.2.1.-); alpha-L-arabinofuranosidase (EC 3.2.1.55); arabinanase (EC 3.2.1.99); xylanase (EC 3.2.1.8); galactan 1,3-beta-galactosidase (EC 3.2.1.145) PF04616 17 Glycosyl hydrolases family 43 CBM4 16 Modules of approx. 150 residues found in bacterial enzymes. Binding of these modules has been demonstrated with xylan, beta-1,3-glucan, beta-1,3-1,4-glucan, beta-1,6-glucan and amorphous cellulose but not with crystalline cellulose. -

Carbohydrate Analysis: Enzymes, Kits and Reagents
2007 Volume 2 Number 3 FOR LIFE SCIENCE RESEARCH ENZYMES, KITS AND REAGENTS FOR ANALYSIS OF: AGAROSE ALGINIC ACID CELLULOSE, LICHENEN AND GLUCANS HEMICELLULOSE AND XYLAN CHITIN AND CHITOSAN CHONDROITINS DEXTRAN HEPARANS HYALURONIC ACID INULIN PEPTIDOGLYCAN PECTIN Cellulose, one of the most abundant biopolymers on earth, is a linear polymer of β-(1-4)-D-glucopyranosyl units. Inter- and PULLULAN intra-chain hydrogen bonding is shown in red. STARCH AND GLYCOGEN Complex Carbohydrate Analysis: Enzymes, Kits and Reagents sigma-aldrich.com The Online Resource for Nutrition Research Products Only from Sigma-Aldrich esigned to help you locate the chemicals and kits The Bioactive Nutrient Explorer now includes a • New! Search for Plants Dneeded to support your work, the Bioactive searchable database of plants listed by physiological Associated with Nutrient Explorer is an Internet-based tool created activity in key areas of research, such as cancer, dia- Physiological Activity to aid medical researchers, pharmacologists, nutrition betes, metabolism and other disease or normal states. • Locate Chemicals found and animal scientists, and analytical chemists study- Plant Detail pages include common and Latin syn- in Specific Plants ing dietary plants and supplements. onyms and display associated physiological activities, • Identify Structurally The Bioactive Nutrient Explorer identifies the while Product Detail pages show the structure family Related Compounds compounds found in a specific plant and arranges and plants that contain the compound, along with them by chemical family and class. You can also comparative product information for easy selection. search for compounds having a similar chemical When you have found the product you need, a structure or for plants containing a specific simple mouse click connects you to our easy online compound. -

NAPRALERT Classification Codes
NAPRALERT Classification Codes June 1993 STN International® Copyright © 1993 American Chemical Society Quoting or copying of material from this publication for educational purposes is encouraged, providing acknowledgement is made of the source of such material. Classification Codes in NAPRALERT The NAPRALERT File contains classification codes that designate pharmacological activities. The code and a corresponding textual description are searchable in the /CC field. To be comprehensive, both the code and the text should be searched. Either may be posted, but not both. The following tables list the code and the text for the various categories. The first two digits of the code describe the categories. Each table lists the category described by codes. The last table (starting on page 56) lists the Classification Codes alphabetically. The text is followed by the code that also describes the category. General types of pharmacological activities may encompass several different categories of effect. You may want to search several classification codes, depending upon how general or specific you want the retrievals to be. By reading through the list, you may find several categories related to the information of interest to you. For example, if you are looking for information on diabetes, you might want to included both HYPOGLYCEMIC ACTIVITY/CC and ANTIHYPERGLYCEMIC ACTIVITY/CC and their codes in the search profile. Use the EXPAND command to verify search terms. => S HYPOGLYCEMIC ACTIVITY/CC OR 17006/CC OR ANTIHYPERGLYCEMIC ACTIVITY/CC OR 17007/CC 490 “HYPOGLYCEMIC”/CC 26131 “ACTIVITY”/CC 490 HYPOGLYCEMIC ACTIVITY/CC ((“HYPOGLYCEMIC”(S)”ACTIVITY”)/CC) 6 17006/CC 776 “ANTIHYPERGLYCEMIC”/CC 26131 “ACTIVITY”/CC 776 ANTIHYPERGLYCEMIC ACTIVITY/CC ((“ANTIHYPERGLYCEMIC”(S)”ACTIVITY”)/CC) 3 17007/CC L1 1038 HYPOGLYCEMIC ACTIVITY/CC OR 17006/CC OR ANTIHYPERGLYCEMIC ACTIVITY/CC OR 17007/CC 2 This search retrieves records with the searched classification codes such as the ones shown here. -

Kinetic and Stability Improvement of Immobilized Pepper Chitosanase
Saudi Journal of Medical and Pharmaceutical Sciences ISSN 2413-4929 (Print) Scholars Middle East Publishers ISSN 2413-4910 (Online) Dubai, United Arab Emirates Website: http://scholarsmepub.com/ Kinetic and Stability Improvement of Immobilized Pepper Chitosanase on Chitin by Covalent Bond Sanaa T El-Sayed1*, Nagwa I Omar1, El-Sayed M El-Sayed2, Wafaa G Shousha2 1Biochemistry Department, National Research Centre, Giza, Egypt 2Chemistry Department, Faculty of Science, Helwan University, Helwan, Egypt Abstract: Kinetic properties and stability studies of pepper chitosanase immobilized *Corresponding author on chitin by covalent bond for production of chitooligosaccharides was the main Sanaa T El-Sayed objective of this work. A comparison with the properties that shown by the free one was also carried out. Maximum enzyme activity was observed at 2.825 and 0.576 mg Article History chitosan /reaction mixture for free chitosanase (FC) and immobilized chitosanase Received: 23.08.2017 (IC), respectively. They followed Michaelis-Menten Kinetics. The values of the Accepted: 31.09.2017 Michaelis-Menten constant indicated that IC exhibited higher affinity toward chitosan Published: 30.09.2017 than that of the free one. Km value was 35.71 mg/reaction mixture for the IC and 17.86 mg/reaction mixture for the free one. The maximum velocities (Vmax) were 12.5 DOI: and 47.62 U/mg for IC and FC, respectively. The optimum pH of IC was slightly 10.21276/sjmps.2017.3.9.3 shifted to acidic range (pH 5.6) from 5.8, which is the optimum for free one. IC exhibited high activity at wide range of temperature from 40 to 60ºC with optimum at 55ºC, while high activity of FC was found at range from 40 to 55ºC with optimum at 45ºC.