Supplementary Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Causes and Evaluation of Mildly Elevated Liver Transaminase Levels ROBERT C
Causes and Evaluation of Mildly Elevated Liver Transaminase Levels ROBERT C. OH, LTC, MC, USA, and THOMAS R. HUSTEAD, LTC, MC, USA Tripler Army Medical Center Family Medicine Residency Program, Honolulu, Hawaii Mild elevations in levels of the liver enzymes alanine transaminase and aspartate transaminase are commonly dis- covered in asymptomatic patients in primary care. Evidence to guide the diagnostic workup is limited. If the history and physical examination do not suggest a cause, a stepwise evaluation should be initiated based on the prevalence of diseases that cause mild elevations in transaminase levels. The most common cause is nonalcoholic fatty liver disease, which can affect up to 30 percent of the population. Other common causes include alcoholic liver disease, medication- associated liver injury, viral hepatitis (hepatitis B and C), and hemochromatosis. Less common causes include α1-antitrypsin deficiency, autoimmune hepatitis, and Wilson disease. Extrahepatic conditions (e.g., thyroid disorders, celiac disease, hemolysis, muscle disorders) can also cause elevated liver transaminase levels. Initial testing should include a fasting lipid profile; measurement of glucose, serum iron, and ferritin; total iron-binding capacity; and hepa- titis B surface antigen and hepatitis C virus antibody testing. If test results are normal, a trial of lifestyle modification with observation or further testing for less common causes is appropriate. Additional testing may include ultrasonog- raphy; measurement of α1-antitrypsin and ceruloplasmin; serum protein electrophoresis; and antinuclear antibody, smooth muscle antibody, and liver/kidney microsomal antibody type 1 testing. Referral for further evaluation and possible liver biopsy is recommended if transaminase levels remain elevated for six months or more. -

Structural Insights Into Histone Modifying Enzymes
Wayne State University Wayne State University Theses January 2019 Structural Insights Into Histone Modifying Enzymes Shruti Amle Wayne State University, [email protected] Follow this and additional works at: https://digitalcommons.wayne.edu/oa_theses Part of the Biochemistry Commons, and the Molecular Biology Commons Recommended Citation Amle, Shruti, "Structural Insights Into Histone Modifying Enzymes" (2019). Wayne State University Theses. 693. https://digitalcommons.wayne.edu/oa_theses/693 This Open Access Embargo is brought to you for free and open access by DigitalCommons@WayneState. It has been accepted for inclusion in Wayne State University Theses by an authorized administrator of DigitalCommons@WayneState. STRUCTURAL INSIGHTS INTO HISTONE MODIFYING ENZYMES by SHRUTI AMLE THESIS Submitted to the Graduate School of Wayne State University, Detroit, Michigan in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE 2019 MAJOR: BIOCHEMISTRY AND MOLECULAR BIOLOGY Approved By: _________________________________________ Advisor Date _________________________________________ _________________________________________ _________________________________________ i ACKNOWLEDGEMENTS Writing this thesis has been extremely captivating and gratifying. I take this opportunity to express my deep and sincere acknowledgements to the number of people for extending their generous support and unstinted help during my entire study. Firstly, I would like to express my respectful regards and deep sense of gratitude to my advisor, Dr. Zhe Yang. I am extremely honored to study and work under his guidance. His vision, ideals, timely motivation and immense knowledge had a deep influence on my entire journey of this career. Without his understanding and support, it would not have been possible to complete this research successfully. I also owe my special thanks to my committee members: Dr. -

Neuroleptic Malignant Syndrome: Another Medical Cause of Acute Abdomen T.C.N
Postgraduate Medical Journal (1989) 65, 653 - 655 Postgrad Med J: first published as 10.1136/pgmj.65.767.653 on 1 September 1989. Downloaded from Missed Diagnosis Neuroleptic malignant syndrome: another medical cause of acute abdomen T.C.N. Lo, M.R. Unwin and I.W. Dymock Department ofMedicine, Stepping Hill Hospital, Stockport, UK. Summary: We present a patient with neuroleptic malignant syndrome and intestinal pseudo- obstruction misdiagnosed as being secondary to septicaemia. The management of the patient is discussed with emphasis on the role of creatine kinase and liver function tests. Introduction Neuroleptic malignant syndrome (NMS) is an occas- (92% neutrophils), serum sodium 130 mmol/l, potas- ional but potentially lethal idiosyncratic complication sium 5.1 mmol/l, urea 8.8 mmol/l, creatinine 79 1tmol/ ofneuroleptic drugs.'"2 By February 1989 the Commit- 1, bilirubin 15 Amol/l, alanine transaminase 837 IU/i tee on Safety of Medicines had received reports of 99 (normal 7-45), aspartate transaminase 392 IU/l (nor- cases. (Committee on Safety of Medicines, personal mal 9-41), gamma glutamyl transferase 15 IU/I (nor- Protected by copyright. Communication). It is thought that the condition is mal <65), alkaline phosphatase 62 IU/I (normal underdiagnosed.34 We report on a case of NMS of 35-125). Serial electrocardiograms showed sinus which the presenting features and therapeutic comp- tachycardia with no acute change. Abdominal X-ray lications occurring during the course of the illness revealed marked gaseous distension ofsmall and large served to further our knowledge in this condition. bowels with multiple fluid levels seen on decubitus films. -

Polymerase Ribozyme with Promoter Recognition
In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition by Razvan Cojocaru BSc, Simon Fraser University, 2014 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Department of Molecular Biology and Biochemistry Faculty of Science © Razvan Cojocaru 2021 SIMON FRASER UNIVERSITY Summer 2021 Copyright in this work is held by the author. Please ensure that any reproduction or re-use is done in accordance with the relevant national copyright legislation. Declaration of Committee Name: Razvan Cojocaru Degree: Doctor of Philosophy Title: In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition Committee: Chair: Lisa Craig Professor, Molecular Biology and Biochemistry Peter Unrau Supervisor Professor, Molecular Biology and Biochemistry Dipankar Sen Committee Member Professor, Molecular Biology and Biochemistry Michel Leroux Committee Member Professor, Molecular Biology and Biochemistry Mani Larijani Internal Examiner Associate Professor, Molecular Biology and Biochemistry Gerald Joyce External Examiner Professor, Jack H. Skirball Center for Chemical Biology and Proteomics Salk Institute for Biological Studies Date Defended/Approved: August 12, 2021 ii Abstract The RNA World hypothesis proposes that the early evolution of life began with RNAs that can serve both as carriers of genetic information and as catalysts. Later in evolution, these functions were gradually replaced by DNA and enzymatic proteins in cellular biology. I start by reviewing the naturally occurring catalytic RNAs, ribozymes, as they play many important roles in biology today. These ribozymes are central to protein synthesis and the regulation of gene expression, creating a landscape that strongly supports an early RNA World. -

Structural Properties of the Nickel Ions in Urease: Novel Insights Into the Catalytic and Inhibition Mechanisms
Coordination Chemistry Reviews 190–192 (1999) 331–355 www.elsevier.com/locate/ccr Structural properties of the nickel ions in urease: novel insights into the catalytic and inhibition mechanisms Stefano Ciurli a,*, Stefano Benini b, Wojciech R. Rypniewski b, Keith S. Wilson c, Silvia Miletti a, Stefano Mangani d a Institute of Agricultural Chemistry, Uni6ersity of Bologna, Viale Berti Pichat 10, I-40127 Bologna, Italy b European Molecular Biology Laboratory, c/o DESY, Notkestraße 85, D-22603 Hamburg, Germany c Department of Chemistry, Uni6ersity of York, Heslington, York YO15DD, UK d Department of Chemistry, Uni6ersity of Siena, Pian dei Mantellini 44, I-53100 Siena, Italy Accepted 13 March 1999 Contents Abstract.................................................... 331 1. Biological background ......................................... 332 2. Spectroscopic investigations of the urease active site structure .................. 333 3. Crystallographic studies of the native enzyme ............................ 334 4. Crystallographic studies of urease mutants.............................. 341 5. Crystallographic studies of urease–inhibitor complexes ...................... 345 6. Crystallographic study of a transition state analogue bound to urease.............. 348 7. A novel proposal for the urease mechanism ............................. 350 References .................................................. 353 Abstract This work provides a comprehensive critical summary of urease spectroscopy, crystallogra- phy, inhibitor binding, and site-directed -
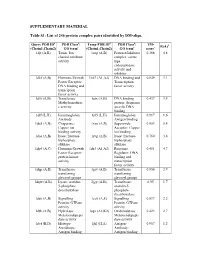
List of 246 Protein Complex Pairs Identified by DM-Align
SUPPLEMENTARY MATERIAL Table S1: List of 246 protein complex pairs identified by DM-align. Query PDB IDa PDB Classb: Temp PDB IDd PDB Classb: TM- R(Å)f (Chain1,Chain2) GO termc (Chain1,Chain2) GO termc scoree 1djt (A,B) Toxin: Ion 1aap (A,B) Protease/Inhibitor 0.368 4.8 channel inhibitor complex: serine activity type endopeptidase activity and inhibitor 1dkf (A,B) Hormone/Growth 1xb7 (A1,A2) DNA binding and 0.849 3.1 Factor Receptor: Transcription DNA binding and factor activity transciption factor activity 1dl5 (A,B) Transferase: 1utx (A,B) DNA binding 0.437 3.9 Methyltransferas protein: Sequence e activity specific DNA binding 1dlf (L,H) Immunoglobin: 1j05 (L,H) Immunoglobin: 0.917 1.6 Antibody Antigen binding 1do5 (A,B) Chaperone: 1xso (A,B) Superoxide 0.953 0.9 Copper ion Acceptor: Copper binding activity ion binding 1dos (A,B) lyase: fructose- 1rvg (A,B) lyase: fructose- 0.760 3.6 biphosphate biphosphate aldolase aldolase 1dp4 (A,C) Hormone/Growth 1dz3 (A1,A2) Response 0.481 4.7 Factor Receptor: Regulator: DNA protein kinase binding and activity transcription factor activity 1dqp (A,B) Transferase: 1grv (A,B) Transferase: 0.856 2.9 transferring transferring glycosyl groups glycosyl groups 1dqw (A,B) Lyase: orotidin- 2jgy (A,B) Transferase: 0.95 1.7 5-phosphate orotidin-5- decarboxylase phosphate decarboxylase 1ds6 (A,B) Signalling 1cc0 (A,E) Signalling 0.897 2.2 Protein: GTPase Protein: GTPase activity activity 1dth (A,B) Hydrolase: 1sqv (A2,K2) Oxidoredutase: 0.423 2.7 Metaloendopepti Metaloendopepti dase activity dase activity 1dvf -

Liver Intracellular L-Cysteine Concentration Is Maintained After Inhibition of the Trans-Sulfuration Pathway by Propargylglycine in Rats
Downloaded from British Journal of Nutrition (1997), 78, 823-831 823 https://www.cambridge.org/core Liver intracellular L-cysteine concentration is maintained after inhibition of the trans-sulfuration pathway by propargylglycine in rats BY ANA TRIGUERO', TERESA BARBER', CONCHA GARC~A', . IP address: INMACULADA R. PUERTES', JUAN SASTRE~AND JUAN R. VIRA' 'Departamento de Bioquhica y Biologia Molecular and 2Departamento de Fisiologfa, Facultades de Farmucia y Medicina, Universidad de Valencia, 46010-Valencia, Spain 170.106.33.14 (Received 31 January 1997 - Revised 24 April I997 - Accepted 7 May 1997) , on 26 Sep 2021 at 14:28:26 To study the fate of L-cysteine and amino acid homeostasis in liver after the inhibition of the trans- sulfuration pathway, rats were treated with propargylglycine (PPG). At 4 h after the administration of PPG, liver cystathionase (EC 4.4.1.1) activity was undetectable, L-cystathionine levels were significantly higher, L-cysteine was unchanged and GSH concentration was significantly lower than values found in livers from control rats injected intraperitoneally with 0.15 M-NaCl. The hepatic levels of amino acids that are intermediates of the urea cycle, L-ornithine, L-citrulline and L-arginine and blood urea were significantly greater. Urea excretion was also higher in PPG-treated rats when , subject to the Cambridge Core terms of use, available at compared with control rats. These data suggest a stimulation of ureagenesis in PPG-treated rats. The inhibition of y-cystathionase was reflected in the blood levels of amino acids, because the L- methionine :L-cyst(e)ine ratio was significantly higher in PPG-treated rats than in control rats; blood concentration of cystathionine was also greater. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Curriculum Vitae Vern Lee Schramm
September 2011 CURRICULUM VITAE VERN LEE SCHRAMM Department of Biochemistry Albert Einstein College of Medicine of Yeshiva University 1300 Morris Park Avenue Bronx, New York 10461 Phone: (718) 430-2813 Fax: (718) 430-8565 E-mail: [email protected] Personal Information: Date of Birth: November 9, 1941 Place of Birth: Howard, South Dakota Citizenship: U.S.A. Home Address: 68 Hampton Oval New Rochelle, NY 10805 Home Telephone: (914) 576-2578 Education: Sept 1959 – June 1963 B.S. in Bacteriology (chemistry emphasis), South Dakota State College Sept 1963 – June 1965 Masters Degree in Nutrition (biochemistry emphasis), Harvard University Research Advisor, Dr. R.P. Geyer Oct 1965 – April 1969 Ph.D. in Mechanism of Enzyme Action, Department of Biochemistry, Australian National University Research Advisor, Dr. John Morrison Postdoctoral Experience: Aug 1969 – Aug 1971 NRC-NSF Postdoctoral Research Associate at NASA Ames Research Center, Biological Adaptation Branch Appointments: July 1999 – Present University Professor of the Albert Einstein College of Medicine July 1995 – Present Ruth Merns Endowed Chair of Biochemistry Aug 1987 – Present Professor and Chairman, Department of Biochemistry, Albert Einstein College of Medicine July 1981 - July 1987 Professor of Biochemistry, Temple University School of Medicine July 1976 - June 1981 Associate Professor of Biochemistry, Temple University School of Medicine Aug 1971 - July 1976 Assistant Professor of Biochemistry, Temple University School of Medicine Vern L. Schramm 2 Fields of Interest: Enzymatic -

Autodisplay: One-Component System for Efficient Surface Display And
JOURNAL OF BACTERIOLOGY, Feb. 1997, p. 794–804 Vol. 179, No. 3 0021-9193/97/$04.0010 Copyright q 1997, American Society for Microbiology Autodisplay: One-Component System for Efficient Surface Display and Release of Soluble Recombinant Proteins from Escherichia coli 1 2 1,2 JOCHEN MAURER, JOACHIM JOSE, AND THOMAS F. MEYER * Downloaded from Abteilung Infektionsbiologie, Max-Planck-Institut fu¨r Biologie, 72076 Tu¨bingen,1 and Abteilung Molekulare Biologie, Max-Planck-Institut fu¨r Infektionsbiologie, 10117 Berlin,2 Germany Received 26 August 1996/Accepted 24 September 1996 The immunoglobulin A protease family of secreted proteins are derived from self-translocating polypro- tein precursors which contain C-terminal domains promoting the translocation of the N-terminally attached passenger domains across gram-negative bacterial outer membranes. Computer predictions identified the C-terminal domain of the Escherichia coli adhesin involved in diffuse adherence (AIDA-I) as a member of the autotransporter family. A model of the b-barrel structure, proposed to be responsible for http://jb.asm.org/ outer membrane translocation, served as a basis for the construction of fusion proteins containing heterologous passengers. Autotransporter-mediated surface display (autodisplay) was investigated for the cholera toxin B subunit and the peptide antigen tag PEYFK. Up to 5% of total cellular protein was detectable in the outer membrane as passenger autotransporter fusion protein synthesized under control of the constitutive PTK promoter. Efficient presentation of the passenger domains was demonstrated in the outer membrane protease T-deficient (ompT) strain E. coli UT5600 and the ompT dsbA double mutant JK321. Surface exposure was ascertained by enzyme-linked immunosorbent assay, immunofluorescence microscopy, and immunogold electron microscopy using antisera specific for the passenger domains. -

The Structure of Allophanate Hydrolase from Granulibacter Bethesdensis Provides Insights Into Substrate Specificity in the Amidase Signature Family
Marquette University e-Publications@Marquette Biological Sciences Faculty Research and Publications Biological Sciences, Department of 2013 The Structure of Allophanate Hydrolase from Granulibacter bethesdensis Provides Insights into Substrate Specificity in the Amidase Signature Family Yi Lin Marquette University, [email protected] Martin St. Maurice Marquette University, [email protected] Follow this and additional works at: https://epublications.marquette.edu/bio_fac Part of the Biochemistry, Biophysics, and Structural Biology Commons Recommended Citation Lin, Yi and St. Maurice, Martin, "The Structure of Allophanate Hydrolase from Granulibacter bethesdensis Provides Insights into Substrate Specificity in the Amidase Signature Family" (2013). Biological Sciences Faculty Research and Publications. 138. https://epublications.marquette.edu/bio_fac/138 Marquette University e-Publications@Marquette Biological Sciences Faculty Research and Publications/College of Arts and Sciences This paper is NOT THE PUBLISHED VERSION; but the author’s final, peer-reviewed manuscript. The published version may be accessed by following the link in the citation below. Biochemistry, Vol. 54, No. 4 (January 29, 2013): 690-700. DOI. This article is © American Chemical Society Publications and permission has been granted for this version to appear in e- Publications@Marquette. American Chemical Society Publications does not grant permission for this article to be further copied/distributed or hosted elsewhere without the express permission from American Chemical Society Publications. The Structure of Allophanate Hydrolase from Granulibacter bethesdensis Provides Insights into Substrate Specificity in the Amidase Signature Family Yi Lin Department of Biological Sciences, Marquette University, Milwaukee, WI Martin St. Maurice Department of Biological Sciences, Marquette University, Milwaukee, WI Abstract Allophanate hydrolase (AH) catalyzes the hydrolysis of allophanate, an intermediate in atrazine degradation and urea catabolism pathways, to NH3 and CO2. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase