Grasnotice 946, Lactobacillus Plantarum Strain DSM 33452
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Effect of Selected Herbal Extracts on Lactic Acid Bacteria Activity
applied sciences Article The Effect of Selected Herbal Extracts on Lactic Acid Bacteria Activity Małgorzata Ziarno 1,* , Mariola Kozłowska 2 , Iwona Scibisz´ 3 , Mariusz Kowalczyk 4 , Sylwia Pawelec 4 , Anna Stochmal 4 and Bartłomiej Szleszy ´nski 5 1 Division of Milk Technology, Department of Food Technology and Assessment, Institute of Food Science, Warsaw University of Life Sciences–SGGW (WULS–SGGW), 02-787 Warsaw, Poland 2 Department of Chemistry, Institute of Food Science, Warsaw University of Life Sciences–SGGW (WULS–SGGW), 02-787 Warsaw, Poland; [email protected] 3 Division of Fruit, Vegetable and Cereal Technology, Department of Food Technology and Assessment, Institute of Food Science, Warsaw University of Life Sciences–SGGW (WULS–SGGW), 02-787 Warsaw, Poland; [email protected] 4 Department of Biochemistry and Crop Quality, Institute of Soil Science and Plant Cultivation, State Research Institute, 24-100 Puławy, Poland; [email protected] (M.K.); [email protected] (S.P.); [email protected] (A.S.) 5 Institute of Horticultural Sciences, Warsaw University of Life Sciences–SGGW (WULS–SGGW), 02-787 Warsaw, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-225-937-666 Abstract: This study aimed to investigate the effect of plant extracts (valerian Valeriana officinalis L., sage Salvia officinalis L., chamomile Matricaria chamomilla L., cistus Cistus L., linden blossom Tilia L., ribwort plantain Plantago lanceolata L., marshmallow Althaea L.) on the activity and growth of lactic acid bacteria (LAB) during the fermentation and passage of milk through a digestive system model. Citation: Ziarno, M.; Kozłowska, M.; The tested extracts were also characterized in terms of their content of polyphenolic compounds and Scibisz,´ I.; Kowalczyk, M.; Pawelec, S.; antioxidant activity. -

A Taxonomic Note on the Genus Lactobacillus
Taxonomic Description template 1 A taxonomic note on the genus Lactobacillus: 2 Description of 23 novel genera, emended description 3 of the genus Lactobacillus Beijerinck 1901, and union 4 of Lactobacillaceae and Leuconostocaceae 5 Jinshui Zheng1, $, Stijn Wittouck2, $, Elisa Salvetti3, $, Charles M.A.P. Franz4, Hugh M.B. Harris5, Paola 6 Mattarelli6, Paul W. O’Toole5, Bruno Pot7, Peter Vandamme8, Jens Walter9, 10, Koichi Watanabe11, 12, 7 Sander Wuyts2, Giovanna E. Felis3, #*, Michael G. Gänzle9, 13#*, Sarah Lebeer2 # 8 '© [Jinshui Zheng, Stijn Wittouck, Elisa Salvetti, Charles M.A.P. Franz, Hugh M.B. Harris, Paola 9 Mattarelli, Paul W. O’Toole, Bruno Pot, Peter Vandamme, Jens Walter, Koichi Watanabe, Sander 10 Wuyts, Giovanna E. Felis, Michael G. Gänzle, Sarah Lebeer]. 11 The definitive peer reviewed, edited version of this article is published in International Journal of 12 Systematic and Evolutionary Microbiology, https://doi.org/10.1099/ijsem.0.004107 13 1Huazhong Agricultural University, State Key Laboratory of Agricultural Microbiology, Hubei Key 14 Laboratory of Agricultural Bioinformatics, Wuhan, Hubei, P.R. China. 15 2Research Group Environmental Ecology and Applied Microbiology, Department of Bioscience 16 Engineering, University of Antwerp, Antwerp, Belgium 17 3 Dept. of Biotechnology, University of Verona, Verona, Italy 18 4 Max Rubner‐Institut, Department of Microbiology and Biotechnology, Kiel, Germany 19 5 School of Microbiology & APC Microbiome Ireland, University College Cork, Co. Cork, Ireland 20 6 University of Bologna, Dept. of Agricultural and Food Sciences, Bologna, Italy 21 7 Research Group of Industrial Microbiology and Food Biotechnology (IMDO), Vrije Universiteit 22 Brussel, Brussels, Belgium 23 8 Laboratory of Microbiology, Department of Biochemistry and Microbiology, Ghent University, Ghent, 24 Belgium 25 9 Department of Agricultural, Food & Nutritional Science, University of Alberta, Edmonton, Canada 26 10 Department of Biological Sciences, University of Alberta, Edmonton, Canada 27 11 National Taiwan University, Dept. -

Download Author Version (PDF)
Environmental Science: Nano CdS Nanoparticles in Soil Induce Metabolic Reprogramming in Broad Bean (Vicia faba L.) Roots and Leaves Journal: Environmental Science: Nano Manuscript ID EN-ART-08-2019-000933.R1 Article Type: Paper Page 1 of 36 Environmental Science: Nano 1 2 3 4 The rapid development of nanotechnology has raised concern regarding the 5 6 7 environmental toxicity of nanoparticles (NPs). However, little is known about the 8 9 molecular mechanisms underlying NP toxicity in plants. Understanding toxic 10 11 12 mechanisms in organisms at molecular level, especially in crop plants, is important for 13 14 their sustainable use and development. In this study, although none of the phenotypic 15 16 17 parameters of broad bean plants (photosynthetic pigments contents, biomass, and lipid 18 19 20 peroxidation) were overtly impacted in response to CdS-NPs in soil during 28 d of 21 22 exposure, metabolomics revealed marked and statistically significant alterations in the 23 24 25 metabolite profiles of plant roots and leaves. The reprogramming of antioxidant 26 27 metabolite production presumably reflected the molecular defense response of the 28 29 30 plants to CdS-NPs stress. The sensitive responses of flavone, putrescine and 31 32 33 noradrenaline in the leaves suggests the use of these compounds in legumes as 34 35 biomarkers of oxidative stress induced by the presence of CdS-NPs in soil. 36 37 38 Metabolomics might thus be a suitable approach for the early detection of soil 39 40 contamination by Cd. In the plants, the reprogramming of carbon and nitrogen 41 42 43 metabolism (including sugars, organic acids, amino acids, and N-containing 44 45 46 compounds) alleviated the toxicity of CdS-NPs, which may have been caused by free 47 48 Cd2+ ions or perhaps by a particle-specific response. -

And Honeydew Sugars with Respect to Their Utilization by the Hymenopteran Parasitoid Cotesia Glomerata F.L
Journal of Insect Physiology 47 (2001) 1077–1084 www.elsevier.com/locate/jinsphys A comparison of nectar- and honeydew sugars with respect to their utilization by the hymenopteran parasitoid Cotesia glomerata F.L. Wa¨ckers * Institute of Plant Sciences, Applied Entomology, Swiss Federal Institute of Technology (ETH), 8092 Zurich, Switzerland Received 10 October 2000; received in revised form 12 February 2001; accepted 19 February 2001 Abstract Fourteen naturally occurring sugars were individually tested with respect to their effect on Cotesia glomerata longevity. Parasitoids kept with solutions of either sucrose, glucose and fructose lived for Ͼ30 days. This constitutes a factor 15 increase in life span in comparison to control individuals kept with water only. Stachyose, mannose, melezitose, melibiose, maltose and erlose increased parasitoid longevity by a factor of 11.2–6.9. Solutions of galactose and trehalose had a marginal, but still significant effect. Lactose and raffinose did not raise parasitoid longevity, while rhamnose actually reduced parasitoid survival. In an additional experiment, the relationship between quantity of sugar consumption and longevity was established for all 14 sugars. To study the effect of an unsuitable sugar in sugar mixtures, a range of glucose:rhamnose mixtures was tested. Even at 20% of the sugar mixture rhamnose suppressed the nutritional benefit of the 80% glucose. The nutritional suitability of the sugars shows a positive correlation with the previously reported gustatory response towards the individual sugars. Patterns of sugar utilization are discussed with respect to hydrolytic enzymes and carbohydrate biochemical characteristics. Our findings for C. glomerata are compared to patterns of sugar utilization reported for other species. -

Electronic Supplementary Information
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2019 Electronic Supplementary Information Poly(ionic liquid)s as a Distinct Receptor Material to Create Highly- Integrated Sensing Platform for Efficiently Identifying a Myriad of Saccharides Wanlin Zhang, Yao Li, Yun Liang, Ning Gao, Chengcheng Liu, Shiqiang Wang, Xianpeng Yin, and Guangtao Li* *Corresponding authors: Guangtao Li ([email protected]) S1 Contents 1. Experimental Section (Page S4-S6) Materials and Characterization (Page S4) Experimental Details (Page S4-S6) 2. Figures and Tables (Page S7-S40) Fig. S1 SEM image of silica colloidal crystal spheres and PIL inverse opal spheres. (Page S7) Fig. S2 Adsorption isotherm of PIL inverse opal. (Page S7) Fig. S3 Dynamic mechanical analysis and thermal gravimetric analysis of PIL materials. (Page S7) Fig. S4 Chemical structures of 23 saccharides. (Page S8) Fig. S5 The counteranion exchange of PIL photonic spheres from Br- to DCA. (Page S9) Fig. S6 Reflection and emission spectra of spheres for saccharides. (Page S9) Table S1 The jack-knifed classification on single-sphere array for 23 saccharides. (Page S10) Fig. S7 Lower detection concentration at 10 mM of the single-sphere array. (Page S11) Fig. S8 Lower detection concentration at 1 mM of the single-sphere array. (Page S12) Fig. S9 PIL sphere exhibiting great pH robustness within the biological pH range. (Page S12) Fig. S10 Exploring the tolerance of PIL spheres to different conditions. (Page S13) Fig. S11 Exploring the reusability of PIL spheres. (Page S14) Fig. S12 Responses of spheres to sugar alcohols. (Page S15) Fig. -
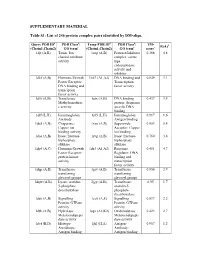
List of 246 Protein Complex Pairs Identified by DM-Align
SUPPLEMENTARY MATERIAL Table S1: List of 246 protein complex pairs identified by DM-align. Query PDB IDa PDB Classb: Temp PDB IDd PDB Classb: TM- R(Å)f (Chain1,Chain2) GO termc (Chain1,Chain2) GO termc scoree 1djt (A,B) Toxin: Ion 1aap (A,B) Protease/Inhibitor 0.368 4.8 channel inhibitor complex: serine activity type endopeptidase activity and inhibitor 1dkf (A,B) Hormone/Growth 1xb7 (A1,A2) DNA binding and 0.849 3.1 Factor Receptor: Transcription DNA binding and factor activity transciption factor activity 1dl5 (A,B) Transferase: 1utx (A,B) DNA binding 0.437 3.9 Methyltransferas protein: Sequence e activity specific DNA binding 1dlf (L,H) Immunoglobin: 1j05 (L,H) Immunoglobin: 0.917 1.6 Antibody Antigen binding 1do5 (A,B) Chaperone: 1xso (A,B) Superoxide 0.953 0.9 Copper ion Acceptor: Copper binding activity ion binding 1dos (A,B) lyase: fructose- 1rvg (A,B) lyase: fructose- 0.760 3.6 biphosphate biphosphate aldolase aldolase 1dp4 (A,C) Hormone/Growth 1dz3 (A1,A2) Response 0.481 4.7 Factor Receptor: Regulator: DNA protein kinase binding and activity transcription factor activity 1dqp (A,B) Transferase: 1grv (A,B) Transferase: 0.856 2.9 transferring transferring glycosyl groups glycosyl groups 1dqw (A,B) Lyase: orotidin- 2jgy (A,B) Transferase: 0.95 1.7 5-phosphate orotidin-5- decarboxylase phosphate decarboxylase 1ds6 (A,B) Signalling 1cc0 (A,E) Signalling 0.897 2.2 Protein: GTPase Protein: GTPase activity activity 1dth (A,B) Hydrolase: 1sqv (A2,K2) Oxidoredutase: 0.423 2.7 Metaloendopepti Metaloendopepti dase activity dase activity 1dvf -

Sugars and Plant Innate Immunity
Journal of Experimental Botany Advance Access published May 2, 2012 Journal of Experimental Botany, Page 1 of 10 doi:10.1093/jxb/ers129 REVIEW PAPER Sugars and plant innate immunity Mohammad Reza Bolouri Moghaddam and Wim Van den Ende* KU Leuven, Laboratory of Molecular Plant Biology, Kasteelpark Arenberg 31, B-3001 Leuven, Belgium * To whom correspondence should be addressed. E-mail: [email protected] Received 27 January 2012; Revised 25 March 2012; Accepted 30 March 2012 Abstract Sugars are involved in many metabolic and signalling pathways in plants. Sugar signals may also contribute to Downloaded from immune responses against pathogens and probably function as priming molecules leading to pathogen-associated molecular patterns (PAMP)-triggered immunity and effector-triggered immunity in plants. These putative roles also depend greatly on coordinated relationships with hormones and the light status in an intricate network. Although evidence in favour of sugar-mediated plant immunity is accumulating, more in-depth fundamental research is required to unravel the sugar signalling pathways involved. This might pave the way for the use of biodegradable http://jxb.oxfordjournals.org/ sugar-(like) compounds to counteract plant diseases as cheaper and safer alternatives for toxic agrochemicals. Key words: immunity, pathogen, priming, signal, sugar. Introduction by guest on March 24, 2016 Sugars such as glucose, fructose, and sucrose are recognized Chisholm et al., 2006). Sugars are well known to activate as signalling molecules in plants (Rolland et al., 2006; various pattern recognition genes (Herbers et al., 1996a,b; Bolouri-Moghaddam et al., 2010), in addition to their Johnson and Ryan, 1990). -

Antifungal Activity of Lactobacillus Pentosus ŁOCK 0979 in the Presence of Polyols and Galactosyl-Polyols
Probiotics & Antimicro. Prot. https://doi.org/10.1007/s12602-017-9344-0 Antifungal Activity of Lactobacillus pentosus ŁOCK 0979 in the Presence of Polyols and Galactosyl-Polyols Lidia Lipińska1 & Robert Klewicki2 & Michał Sójka2 & Radosław Bonikowski3 & Dorota Żyżelewicz2 & Krzysztof Kołodziejczyk2 & Elżbieta Klewicka 1 # The Author(s) 2017. This article is an open access publication Abstract The antifungal activity of Lactobacillus pentosus Keywords Antifungal activity . Galactosyl-polyols . ŁOCK 0979 depends both on the culture medium and on the Lactobacillus . Metabolites . Polyols . SEM fungal species. In the control medium, the strain exhibited limited antagonistic activity against indicator food-borne molds and yeasts. However, the supplementation of the bac- Introduction terial culture medium with polyols (erythritol, lactitol, maltitol, mannitol, sorbitol, xylitol) or their galactosyl deriva- Filamentous fungi and yeasts are present in almost all types of tives (gal-erythritol, gal-sorbitol, gal-xylitol) enhanced the an- ecosystems due to their high adaptation ability and low nutri- tifungal properties of Lactobacillus pentosus ŁOCK 0979. Its tional requirements. Filamentous fungi are widespread food metabolites were identified and quantified by enzymatic spoilage microorganisms responsible for significant economic methods, HPLC, UHPLC-MS coupled with QuEChERS, losses in the agri-food industry [6]; they are also a major and GC-MS. The presence of polyols and gal-polyols signif- health concern due to mycotoxin production. The most com- icantly affected the acid metabolite profile of the bacterial mon genera of spoilage fungi include Penicillium, Fusarium, culture supernatant. In addition, lactitol and mannitol were Aspergillus, Cladosporium,andRhizopus [21]. Commercial used by bacteria as alternative carbon sources. A number of foodstuffs are usually protected from such microorganisms by compounds with potential antifungal properties were identi- physical and chemical techniques. -

Sweetness and Sensory Properties of Commercial and Novel Oligosaccharides Of
1 Sweetness and sensory properties of commercial and novel oligosaccharides of 2 prebiotic potential 3 4 Laura Ruiz-Aceitunoa, Oswaldo Hernandez-Hernandeza, Sofia Kolidab, F. Javier 5 Morenoa,* and Lisa Methvenc 6 7 a Institute of Food Science Research, CIAL (CSIC-UAM), Nicolás Cabrera 9, 28049 8 Madrid (Spain) 9 b OptiBiotix Health plc, Innovation Centre, Innovation Way, Heslington, York YO10 10 5DG (UK) 11 c Sensory Science Centre, Department of Food and Nutritional Sciences, The University 12 of Reading, PO Box 226, Whiteknights, Reading RG6 6AP (UK) 13 14 *Corresponding author: [email protected] Tel (+34) 91 0017948 1 15 Abstract 16 This study investigates the sweetness properties and other sensory attributes of ten 17 commercial and four novel prebiotics (4-galactosyl-kojibiose, lactulosucrose, lactosyl- 18 oligofructosides and raffinosyl-oligofructosides) of high degree of purity and assesses the 19 influence of their chemical structure features on sweetness. The impact of the type of 20 glycosidic linkage by testing four sucrose isomers, as well as the monomer composition 21 and degree of polymerization on sweetness properties were determined. Data from the 22 sensory panel combined with principal component analysis (PCA) concludes that chain 23 length was the most relevant factor in determining the sweetness potential of a 24 carbohydrate. Thus, disaccharides had higher sweetness values than trisaccharides which, 25 in turn, exhibited superior sweetness than mixtures of oligosaccharides having DP above 26 3. Furthermore, a weak non-significant trend indicated that the presence of a ketose sugar 27 moiety led to higher sweetness. The novel prebiotics tested in this study had between 18 28 and 25% of relative sweetness, in line with other commercial prebiotics, and samples 29 varied in their extent of off flavour. -

Using MALDI-TOF MS Analysis for Rapid Discrimination of MRSA MLST
Using MALDI-TOF MS analysis for rapid discrimination of MRSA MLST types Jang-Jih Lu1,2,Tsui-Ping Liu1, Shih-Cheng Chang1,2 1Department of Laboratory Medicine, Chang Gung Memorial Hospital, Linkou, Taoyuan, Taiwan 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan Email: [email protected] 73 0:F8 MS Ra w 71 0:F9 MS Ra w 5000 6422.794 6000 Intens.[a.u.] Intens.[a.u.] Introduction 4000 Results ST5 ST5 4000 3000 2978.985 2000 6551.531 Staphylococcus aureus is one of the common causes of 3027.802 3054.785 2964.900 3175.943 2000 Table 1: MRSA and MSSA associated peaks by MALDI TOF 1000 community-associated infections and health care related 234 0:F1 MS Ra w 234 0:F1 MS Ra w 3005.919 MRSA(129 isolates) MSSA(77 isolates) 5000 6000 Intens.[a.u.] Intens.[a.u.] pathogen. It can usually cause wound infection, ST45 4000 ST45 6549.206 4000 3000 MS peaks (m/z) no. of isolate % of isolate no. of isolate % of isolate 6420.525 osteomyelitis, and even serious invasive infections, such as 2000 6610.628 2000 2978.373 3053.663 3174.805 2415 67 51.9 2 2.6 1000 sepsis and pneumonia. Epidemiology studies have shown 86 0:E4 MS Ra w 70 0:E5 MS Ra w 3058.565 5000 2431 65 50.4 0 0 3074.390 6000 Intens.[a.u.] Intens.[a.u.] that there are close relationship between the results of ST59 4000 6550.723 ST59 4000 3000 2879 51 39.5 2 2.6 6422.151 genotyping from multilocus sequence typing (MLST) and 2000 2000 characteristics of MRSA strains, including hospital- 6593 77 59.7 7 9.1 1000 200 0:E8 MS Ra w 38 0:E11 MS Ra w Appear of any above 103 79.8 11 14.3 2978.890 5000 6000 Intens.[a.u.] Intens.[a.u.] 6421.658 acquired, community-associated strain and even the 2965.568 ST239 4000 ST239 peaks 4000 3000 none of above peaks 6590.270 26 20.1 66 85.7 2937.113 3177.333 2916.384 antibiogram of the strain. -

The Impact of Oil Type and Lactic Acid Bacteria on Conjugated Linoleic Acid Production
JOBIMB, 2016, Vol 4, No 2, 25-29 JOURNAL OF BIOCHEMISTRY, MICROBIOLOGY AND BIOTECHNOLOGY Website: http://journal.hibiscuspublisher.com/index.php/JOBIMB/index The Impact of Oil Type and Lactic Acid Bacteria on Conjugated Linoleic Acid Production Mahmoud A. Al-Saman 1*, Rafaat M. Elsanhoty 1 and Elhadary A. E. 2 1Department of Industrial Biotechnology, Genetic Engineering and Biotechnology Research Institute, University of Sadat City, Sadat City 22857/79, Egypt. 2Biochemistry Department, Faculty of Agriculture, Benha University, Egypt. *Corresponding author: Dr. Mahmoud Abd El-Hamid Al-Saman Department of Industrial Biotechnology, Genetic Engineering and Biotechnology Research Institute, University of Sadat City, Sadat City 22857/79, Egypt. Email: [email protected] [email protected] HISTORY ABSTRACT This work was conducted to investigate the effect of oil type and lactic acid bacteria on the Received: 27 th October 2016 conjugated linoleic acid (CLA) production in MRS medium. The ability of eight strains of Received in revised form: 2nd December 2016 Accepted: 17th December 2016 lactic acids bacteria; Lactobacillus acidophilus (P2, ATCC 20552), Lactobacillus brevis (P102), Lactobacillus casei (P9, DSMZ 20011), Lactobacillus plantarum (P1), Lactobacillus KEYWORDS pentosus (P4), Lactobacillus rhamnosus (P5, TISTR 541), Bifidobacterium longum (BL) and conjugated linoleic acid (CLA) Bifidobacterium lactis (P7, Bb-12) for the production of CLA in the MRS broth was lactic acid bacteria investigated. Two vegetable oils (sun flower oil & linseed oil) and cod liver oil were used as vegetable oils cod liver oil substrates in MRS media. The oils were added to MRS in concentration of 10 mg/ml and probiotic incubated for three days at 37°C. -

Multi-Product Lactic Acid Bacteria Fermentations: a Review
fermentation Review Multi-Product Lactic Acid Bacteria Fermentations: A Review José Aníbal Mora-Villalobos 1 ,Jéssica Montero-Zamora 1, Natalia Barboza 2,3, Carolina Rojas-Garbanzo 3, Jessie Usaga 3, Mauricio Redondo-Solano 4, Linda Schroedter 5, Agata Olszewska-Widdrat 5 and José Pablo López-Gómez 5,* 1 National Center for Biotechnological Innovations of Costa Rica (CENIBiot), National Center of High Technology (CeNAT), San Jose 1174-1200, Costa Rica; [email protected] (J.A.M.-V.); [email protected] (J.M.-Z.) 2 Food Technology Department, University of Costa Rica (UCR), San Jose 11501-2060, Costa Rica; [email protected] 3 National Center for Food Science and Technology (CITA), University of Costa Rica (UCR), San Jose 11501-2060, Costa Rica; [email protected] (C.R.-G.); [email protected] (J.U.) 4 Research Center in Tropical Diseases (CIET) and Food Microbiology Section, Microbiology Faculty, University of Costa Rica (UCR), San Jose 11501-2060, Costa Rica; [email protected] 5 Bioengineering Department, Leibniz Institute for Agricultural Engineering and Bioeconomy (ATB), 14469 Potsdam, Germany; [email protected] (L.S.); [email protected] (A.O.-W.) * Correspondence: [email protected]; Tel.: +49-(0331)-5699-857 Received: 15 December 2019; Accepted: 4 February 2020; Published: 10 February 2020 Abstract: Industrial biotechnology is a continuously expanding field focused on the application of microorganisms to produce chemicals using renewable sources as substrates. Currently, an increasing interest in new versatile processes, able to utilize a variety of substrates to obtain diverse products, can be observed.