Consolidated List of Up-Regulated Proteins Expressed at Different Cr (VI) Concentrations at Time Points
Total Page:16
File Type:pdf, Size:1020Kb
Recommended publications
-

Specific Binding of Transposase to Terminal Inverted Repeats Of
Proc. Natl. Acad. Sci. USA Vol. 84, pp. 8220-8224, December 1987 Biochemistry Specific binding of transposase to terminal inverted repeats of transposable element Tn3 (transposon/DNA binding protein/DNase I "footprinting") HITOSHI ICHIKAWA*, KAORU IKEDA*, WILLIAM L. WISHARTt, AND EIICHI OHTSUBO*t *Institute of Applied Microbiology, University of Tokyo, Bunkyo-ku, Yayoi 1-1-1, Tokyo 113, Japan; and tDepartment of Molecular Biology, Princeton University, Princeton, NJ 08544 Communicated by Norman Davidson, July 27, 1987 ABSTRACT Tn3 transposase, which is required for trans- containing the Tn3 terminal regions when 8 mM ATP is position of Tn3, has been purified by a low-ionic-strength- present (14). precipitation method. Using a nitrocellulose filter binding In this paper, we study the interaction of Tn3 transposase, assay, we have shown that transposase binds to any restriction which has been purified by a low-ionic-strength-precipitation fragment. However, binding of the transposase to specific method, with DNA. We demonstrate that the purified fragments containing the terminal inverted repeat sequences of transposase is an ATP-independent DNA binding protein, Tn3 can be demonstrated by treatment of transposase-DNA which has interesting properties that may be involved in the complexes with heparin, which effectively removes the actual transposition event of Tn3. transposase bound to the other nonspecific fragments at pH 5-6. DNase I "footprinting" analysis showed that the MATERIALS AND METHODS transposase protects an inner 25-base-pair region of the 38- base-pair terminal inverted repeat sequence of Tn3. This Bacterial Strains and Plasmids. The bacterial strains used protection is not dependent on pH. -

Polyphosphate-Dependent Synthesis of ATP and ADP by the Family-2 Polyphosphate Kinases in Bacteria
Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria Boguslaw Noceka, Samvel Kochinyanb, Michael Proudfootb, Greg Brownb, Elena Evdokimovaa,b, Jerzy Osipiuka, Aled M. Edwardsa,b, Alexei Savchenkoa,b, Andrzej Joachimiaka,c,1, and Alexander F. Yakuninb,1 aMidwest Center for Structural Genomics and Structural Biology Center, Department of Biosciences, Argonne National Laboratory, 9700 South Cass Avenue, Building 202, Argonne, IL 60439; bBanting and Best Department of Medical Research, University of Toronto, Toronto, ON, Canada M5G 1L6; and cDepartment of Biochemistry and Molecular Biology, University of Chicago, 920 East 58th Street, Chicago, IL 60637 Edited by Robert Haselkorn, University of Chicago, Chicago, IL, and approved September 30, 2008 (received for review August 1, 2008) Inorganic polyphosphate (polyP) is a linear polymer of tens or hun- exopolysaccharide essential for the virulence of P. aeruginosa (12). dreds of phosphate residues linked by high-energy bonds. It is found In the social slime mold Dictyostelium discoideum, a different PPK in all organisms and has been proposed to serve as an energy source was found (DdPPK2) with sequence and properties similar to that in a pre-ATP world. This ubiquitous and abundant biopolymer plays of actin-related proteins (Arps) (14). DdPPK2 is a complex of 3 numerous and vital roles in metabolism and regulation in prokaryotes Arps (Arp1, Arp2, and Arpx) and resembles the actin family in and eukaryotes, but the underlying molecular mechanisms for most molecular weight, sequence, and filamentous structure. Remark- activities of polyP remain unknown. In prokaryotes, the synthesis and ably, DdPPK2 can polymerize into an actin-like filament concurrent utilization of polyP are catalyzed by 2 families of polyP kinases, PPK1 with the synthesis of polyP (14). -

Enzymatic Encoding Methods for Efficient Synthesis Of
(19) TZZ__T (11) EP 1 957 644 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12N 15/10 (2006.01) C12Q 1/68 (2006.01) 01.12.2010 Bulletin 2010/48 C40B 40/06 (2006.01) C40B 50/06 (2006.01) (21) Application number: 06818144.5 (86) International application number: PCT/DK2006/000685 (22) Date of filing: 01.12.2006 (87) International publication number: WO 2007/062664 (07.06.2007 Gazette 2007/23) (54) ENZYMATIC ENCODING METHODS FOR EFFICIENT SYNTHESIS OF LARGE LIBRARIES ENZYMVERMITTELNDE KODIERUNGSMETHODEN FÜR EINE EFFIZIENTE SYNTHESE VON GROSSEN BIBLIOTHEKEN PROCEDES DE CODAGE ENZYMATIQUE DESTINES A LA SYNTHESE EFFICACE DE BIBLIOTHEQUES IMPORTANTES (84) Designated Contracting States: • GOLDBECH, Anne AT BE BG CH CY CZ DE DK EE ES FI FR GB GR DK-2200 Copenhagen N (DK) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • DE LEON, Daen SK TR DK-2300 Copenhagen S (DK) Designated Extension States: • KALDOR, Ditte Kievsmose AL BA HR MK RS DK-2880 Bagsvaerd (DK) • SLØK, Frank Abilgaard (30) Priority: 01.12.2005 DK 200501704 DK-3450 Allerød (DK) 02.12.2005 US 741490 P • HUSEMOEN, Birgitte Nystrup DK-2500 Valby (DK) (43) Date of publication of application: • DOLBERG, Johannes 20.08.2008 Bulletin 2008/34 DK-1674 Copenhagen V (DK) • JENSEN, Kim Birkebæk (73) Proprietor: Nuevolution A/S DK-2610 Rødovre (DK) 2100 Copenhagen 0 (DK) • PETERSEN, Lene DK-2100 Copenhagen Ø (DK) (72) Inventors: • NØRREGAARD-MADSEN, Mads • FRANCH, Thomas DK-3460 Birkerød (DK) DK-3070 Snekkersten (DK) • GODSKESEN, -

A Sleeping Beauty Mutagenesis Screen Reveals a Tumor Suppressor Role for Ncoa2/Src-2 in Liver Cancer
A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer Kathryn A. O’Donnella,b,1,2, Vincent W. Kengc,d,e, Brian Yorkf, Erin L. Reinekef, Daekwan Seog, Danhua Fanc,h, Kevin A. T. Silversteinc,h, Christina T. Schruma,b, Wei Rose Xiea,b,3, Loris Mularonii,j, Sarah J. Wheelani,j, Michael S. Torbensonk, Bert W. O’Malleyf, David A. Largaespadac,d,e, and Jef D. Boekea,b,i,2 Departments of aMolecular Biology and Genetics, iOncology, jDivision of Biostatistics and Bioinformatics, and kPathology, and bThe High Throughput Biology Center, The Johns Hopkins University School of Medicine, Baltimore, MD 21205; cMasonic Cancer Center, dDepartment of Genetics, Cell Biology, and Development, eCenter for Genome Engineering, and hBiostatistics and Bioinformatics Core, University of Minnesota, Minneapolis, MN 55455; fDepartment of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030; and gLaboratory of Experimental Carcinogenesis, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892 AUTHOR SUMMARY Emerging data from cancer ge- functions are altered. This ap- fi nome-sequencing studies have Sleeping Beauty (SB) transposase proach identi ed at least 16 demonstrated that human genes/loci that contribute to liver tumors exhibit tremendous Transposon array with gene trap (GT) tumor development. complexity and heterogeneity in We next validated that the the number and nature of iden- genes identified in the SB screen tified mutations (1). Based on contribute to tumor initiation these findings, there is an in- SB mobilization and/or progression using in vitro creasing need for in vivo vali- and in vivo cancer model sys- dation of genes whose altered tems. -

Phosphatidylinositol-3-Kinase Related Kinases (Pikks) in Radiation-Induced Dna Damage
Mil. Med. Sci. Lett. (Voj. Zdrav. Listy) 2012, vol. 81(4), p. 177-187 ISSN 0372-7025 DOI: 10.31482/mmsl.2012.025 REVIEW ARTICLE PHOSPHATIDYLINOSITOL-3-KINASE RELATED KINASES (PIKKS) IN RADIATION-INDUCED DNA DAMAGE Ales Tichy 1, Kamila Durisova 1, Eva Novotna 1, Lenka Zarybnicka 1, Jirina Vavrova 1, Jaroslav Pejchal 2, Zuzana Sinkorova 1 1 Department of Radiobiology, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic 2 Centrum of Advanced Studies, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic. Received 5 th September 2012. Revised 27 th November 2012. Published 7 th December 2012. Summary This review describes a drug target for cancer therapy, family of phosphatidylinositol-3 kinase related kinases (PIKKs), and it gives a comprehensive review of recent information. Besides general information about phosphatidylinositol-3 kinase superfamily, it characterizes a DNA-damage response pathway since it is monitored by PIKKs. Key words: PIKKs; ATM; ATR; DNA-PK; Ionising radiation; DNA-repair ABBREVIATIONS therapy and radiation play a pivotal role. Since cancer is one of the leading causes of death worldwide, it is DSB - double stand breaks, reasonable to invest time and resources in the enligh - IR - ionising radiation, tening of mechanisms, which underlie radio-resis - p53 - TP53 tumour suppressors, tance. PI - phosphatidylinositol. The aim of this review is to describe the family INTRODUCTION of phosphatidyinositol 3-kinases (PI3K) and its func - tional subgroup - phosphatidylinositol-3-kinase rela - An efficient cancer treatment means to restore ted kinases (PIKKs) and their relation to repairing of controlled tissue growth via interfering with cell sig - radiation-induced DNA damage. -
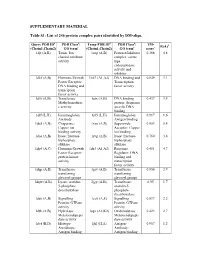
List of 246 Protein Complex Pairs Identified by DM-Align
SUPPLEMENTARY MATERIAL Table S1: List of 246 protein complex pairs identified by DM-align. Query PDB IDa PDB Classb: Temp PDB IDd PDB Classb: TM- R(Å)f (Chain1,Chain2) GO termc (Chain1,Chain2) GO termc scoree 1djt (A,B) Toxin: Ion 1aap (A,B) Protease/Inhibitor 0.368 4.8 channel inhibitor complex: serine activity type endopeptidase activity and inhibitor 1dkf (A,B) Hormone/Growth 1xb7 (A1,A2) DNA binding and 0.849 3.1 Factor Receptor: Transcription DNA binding and factor activity transciption factor activity 1dl5 (A,B) Transferase: 1utx (A,B) DNA binding 0.437 3.9 Methyltransferas protein: Sequence e activity specific DNA binding 1dlf (L,H) Immunoglobin: 1j05 (L,H) Immunoglobin: 0.917 1.6 Antibody Antigen binding 1do5 (A,B) Chaperone: 1xso (A,B) Superoxide 0.953 0.9 Copper ion Acceptor: Copper binding activity ion binding 1dos (A,B) lyase: fructose- 1rvg (A,B) lyase: fructose- 0.760 3.6 biphosphate biphosphate aldolase aldolase 1dp4 (A,C) Hormone/Growth 1dz3 (A1,A2) Response 0.481 4.7 Factor Receptor: Regulator: DNA protein kinase binding and activity transcription factor activity 1dqp (A,B) Transferase: 1grv (A,B) Transferase: 0.856 2.9 transferring transferring glycosyl groups glycosyl groups 1dqw (A,B) Lyase: orotidin- 2jgy (A,B) Transferase: 0.95 1.7 5-phosphate orotidin-5- decarboxylase phosphate decarboxylase 1ds6 (A,B) Signalling 1cc0 (A,E) Signalling 0.897 2.2 Protein: GTPase Protein: GTPase activity activity 1dth (A,B) Hydrolase: 1sqv (A2,K2) Oxidoredutase: 0.423 2.7 Metaloendopepti Metaloendopepti dase activity dase activity 1dvf -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Allosteric Integrase Inhibitor Potency Is Determined Through the Inhibition of HIV-1 Particle Maturation
Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation Kellie A. Juradoa, Hao Wanga, Alison Slaughterb, Lei Fengb, Jacques J. Kesslb, Yasuhiro Koha, Weifeng Wanga, Allison Ballandras-Colasa, Pratiq A. Patelc, James R. Fuchsc, Mamuka Kvaratskheliab, and Alan Engelmana,1 aDepartment of Cancer Immunology and AIDS, Dana-Farber Cancer Institute and Department of Medicine, Harvard Medical School, Boston, MA 02215; and bCenter for Retrovirus Research and Comprehensive Cancer Center and cDivision of Medicinal Chemistry and Pharmacognosy, College of Pharmacy, The Ohio State University, Columbus, OH 43210 Edited by Alan R. Rein, National Cancer Institute, Frederick, MD, and accepted by the Editorial Board April 1, 2013 (received for review January 14, 2013) Integration is essential for HIV-1 replication, and the viral integrase HIV-1 preferentially integrates along the bodies of active genes (IN) protein is an important therapeutic target. Allosteric IN inhib- (6), a trait that is largely attributable to an interaction between itors (ALLINIs) that engage the IN dimer interface at the binding site IN and the host protein lens epithelium-derived growth factor for the host protein lens epithelium-derived growth factor (LEDGF)/ (LEDGF)/transcriptional coactivator p75 (reviewed in refs. 7 and transcriptional coactivator p75 are an emerging class of small mole- 8). LEDGF/p75 functions as a bimodal tether during integration: cule antagonists. Consistent with the inhibition of a multivalent drug elements within its N-terminal region confer constitutive binding to target, ALLINIs display steep antiviral dose–response curves ex vivo. chromatin, whereas a downstream IN-binding domain (IBD) binds ALLINIs multimerize IN protein and concordantly block its assembly lentiviral IN proteins (9, 10). -

Table S1. List of Oligonucleotide Primers Used
Table S1. List of oligonucleotide primers used. Cla4 LF-5' GTAGGATCCGCTCTGTCAAGCCTCCGACC M629Arev CCTCCCTCCATGTACTCcgcGATGACCCAgAGCTCGTTG M629Afwd CAACGAGCTcTGGGTCATCgcgGAGTACATGGAGGGAGG LF-3' GTAGGCCATCTAGGCCGCAATCTCGTCAAGTAAAGTCG RF-5' GTAGGCCTGAGTGGCCCGAGATTGCAACGTGTAACC RF-3' GTAGGATCCCGTACGCTGCGATCGCTTGC Ukc1 LF-5' GCAATATTATGTCTACTTTGAGCG M398Arev CCGCCGGGCAAgAAtTCcgcGAGAAGGTACAGATACGc M398Afwd gCGTATCTGTACCTTCTCgcgGAaTTcTTGCCCGGCGG LF-3' GAGGCCATCTAGGCCATTTACGATGGCAGACAAAGG RF-5' GTGGCCTGAGTGGCCATTGGTTTGGGCGAATGGC RF-3' GCAATATTCGTACGTCAACAGCGCG Nrc2 LF-5' GCAATATTTCGAAAAGGGTCGTTCC M454Grev GCCACCCATGCAGTAcTCgccGCAGAGGTAGAGGTAATC M454Gfwd GATTACCTCTACCTCTGCggcGAgTACTGCATGGGTGGC LF-3' GAGGCCATCTAGGCCGACGAGTGAAGCTTTCGAGCG RF-5' GAGGCCTGAGTGGCCTAAGCATCTTGGCTTCTGC RF-3' GCAATATTCGGTCAACGCTTTTCAGATACC Ipl1 LF-5' GTCAATATTCTACTTTGTGAAGACGCTGC M629Arev GCTCCCCACGACCAGCgAATTCGATagcGAGGAAGACTCGGCCCTCATC M629Afwd GATGAGGGCCGAGTCTTCCTCgctATCGAATTcGCTGGTCGTGGGGAGC LF-3' TGAGGCCATCTAGGCCGGTGCCTTAGATTCCGTATAGC RF-5' CATGGCCTGAGTGGCCGATTCTTCTTCTGTCATCGAC RF-3' GACAATATTGCTGACCTTGTCTACTTGG Ire1 LF-5' GCAATATTAAAGCACAACTCAACGC D1014Arev CCGTAGCCAAGCACCTCGgCCGAtATcGTGAGCGAAG D1014Afwd CTTCGCTCACgATaTCGGcCGAGGTGCTTGGCTACGG LF-3' GAGGCCATCTAGGCCAACTGGGCAAAGGAGATGGA RF-5' GAGGCCTGAGTGGCCGTGCGCCTGTGTATCTCTTTG RF-3' GCAATATTGGCCATCTGAGGGCTGAC Kin28 LF-5' GACAATATTCATCTTTCACCCTTCCAAAG L94Arev TGATGAGTGCTTCTAGATTGGTGTCggcGAAcTCgAGCACCAGGTTG L94Afwd CAACCTGGTGCTcGAgTTCgccGACACCAATCTAGAAGCACTCATCA LF-3' TGAGGCCATCTAGGCCCACAGAGATCCGCTTTAATGC RF-5' CATGGCCTGAGTGGCCAGGGCTAGTACGACCTCG -

Using MALDI-TOF MS Analysis for Rapid Discrimination of MRSA MLST
Using MALDI-TOF MS analysis for rapid discrimination of MRSA MLST types Jang-Jih Lu1,2,Tsui-Ping Liu1, Shih-Cheng Chang1,2 1Department of Laboratory Medicine, Chang Gung Memorial Hospital, Linkou, Taoyuan, Taiwan 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan Email: [email protected] 73 0:F8 MS Ra w 71 0:F9 MS Ra w 5000 6422.794 6000 Intens.[a.u.] Intens.[a.u.] Introduction 4000 Results ST5 ST5 4000 3000 2978.985 2000 6551.531 Staphylococcus aureus is one of the common causes of 3027.802 3054.785 2964.900 3175.943 2000 Table 1: MRSA and MSSA associated peaks by MALDI TOF 1000 community-associated infections and health care related 234 0:F1 MS Ra w 234 0:F1 MS Ra w 3005.919 MRSA(129 isolates) MSSA(77 isolates) 5000 6000 Intens.[a.u.] Intens.[a.u.] pathogen. It can usually cause wound infection, ST45 4000 ST45 6549.206 4000 3000 MS peaks (m/z) no. of isolate % of isolate no. of isolate % of isolate 6420.525 osteomyelitis, and even serious invasive infections, such as 2000 6610.628 2000 2978.373 3053.663 3174.805 2415 67 51.9 2 2.6 1000 sepsis and pneumonia. Epidemiology studies have shown 86 0:E4 MS Ra w 70 0:E5 MS Ra w 3058.565 5000 2431 65 50.4 0 0 3074.390 6000 Intens.[a.u.] Intens.[a.u.] that there are close relationship between the results of ST59 4000 6550.723 ST59 4000 3000 2879 51 39.5 2 2.6 6422.151 genotyping from multilocus sequence typing (MLST) and 2000 2000 characteristics of MRSA strains, including hospital- 6593 77 59.7 7 9.1 1000 200 0:E8 MS Ra w 38 0:E11 MS Ra w Appear of any above 103 79.8 11 14.3 2978.890 5000 6000 Intens.[a.u.] Intens.[a.u.] 6421.658 acquired, community-associated strain and even the 2965.568 ST239 4000 ST239 peaks 4000 3000 none of above peaks 6590.270 26 20.1 66 85.7 2937.113 3177.333 2916.384 antibiogram of the strain. -

ORGANIC CHEMICAL TOXICOLOGY of FISHES This Is Volume 33 in The
ORGANIC CHEMICAL TOXICOLOGY OF FISHES This is Volume 33 in the FISH PHYSIOLOGY series Edited by Anthony P. Farrell and Colin J. Brauner Honorary Editors: William S. Hoar and David J. Randall A complete list of books in this series appears at the end of the volume ORGANIC CHEMICAL TOXICOLOGY OF FISHES Edited by KEITH B. TIERNEY Department of Biological Sciences University of Alberta Edmonton, Alberta Canada ANTHONY P. FARRELL Department of Zoology, and Faculty of Land and Food Systems The University of British Columbia Vancouver, British Columbia Canada COLIN J. BRAUNER Department of Zoology The University of British Columbia Vancouver, British Columbia Canada AMSTERDAM BOSTON HEIDELBERG LONDON NEW YORK OXFORD PARIS SAN DIEGO SAN FRANCISCO SINGAPORE SYDNEY TOKYO Academic Press is an imprint of Elsevier Academic Press is an imprint of Elsevier 32 Jamestown Road, London NW1 7BY, UK 225 Wyman Street, Waltham, MA 02451, USA 525 B Street, Suite 1800, San Diego, CA 92101-4495, USA Copyright r 2014 Elsevier Inc. All rights reserved The cover illustrates the diversity of effects an example synthetic organic water pollutant can have on fish. The chemical shown is 2,4-D, an herbicide that can be found in streams near urbanization and agriculture. The fish shown is one that can live in such streams: rainbow trout (Oncorhynchus mykiss). The effect shown on the left is the ability of 2,4-D (yellow line) to stimulate olfactory sensory neurons vs. control (black line) (measured as an electro- olfactogram; EOG). The effect shown on the right is the ability of 2,4-D to induce the expression of an egg yolk precursor protein (vitellogenin) in male fish. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase