Assay for Detecting Mycoplasma by Measuring Acetate Kinase Or Carbamate Kinase Activity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Katalog 2015 Cover Paul Lin *Hinweis Förderung.Indd
Product List 2015 WE LIVE SERVICE Certificates quartett owns two productions sites that are certified according to EN ISO 9001:2008 Quality management systems - Requirements EN ISO 13485:2012 + AC:2012 Medical devices - Quality management systems - Requirements for regulatory purposes GMP Conformity Our quality management guarantees products of highest quality! 2 Foreword to the quartett product list 2015 quartett Immunodiagnostika, Biotechnologie + Kosmetik Vertriebs GmbH welcomes you as one of our new business partners as well as all of our previous loyal clients. You are now member of quartett´s worldwide customers. First of all we would like to introduce ourselves to you. Founded as a family-run company in 1986, quartett ensures for more than a quarter of a century consistent quality of products. Service and support of our valued customers are our daily businesses. And we will continue! In the end 80´s quartett offered radioimmunoassay and enzyme immunoassay kits from different manufacturers in the USA. In the beginning 90´s the company changed its strategy from offering products for routine diagnostic to the increasing field of research and development. Setting up a production plant in 1997 and a second one in 2011 supported this decision. The company specialized its product profile in the field of manufacturing synthetic peptides for antibody production, peptides such as protease inhibitors, biochemical reagents and products for histology, cytology and immunohistology. All products are exclusively manufactured in Germany without outsourcing any production step. Nowadays, we expand into all other diagnostic and research fields and supply our customers in universities, government institutes, pharmaceutical and biotechnological companies, hospitals, and private doctor offices. -

Recombinant Escheichia Coli-Catalyzed Production of Cytidine 5′-Triphosphate from Cytidine 5′-Monophosphate
J. Ind. Eng. Chem., Vol. 12, No. 5, (2006) 757-761 Recombinant Escheichia coli-Catalyzed Production of Cytidine 5′-Triphosphate from Cytidine 5′-Monophosphate Sun-Gu Lee† and Byung-Gee Kim* Department of Chemical and Biochemical Engineering, Pusan National University, Busan 609-735, Korea *School of Chemical Engineering, and Institute of Molecular Biology and Genetics, Seoul National University, Seoul 151-742, Korea Received April 24, 2006; Accepted May 22, 2006 Abstract: A recombinant Escherichia coli overexpressing CMP-kinase was constructed and employed as a whole cell biocatalyst for the conversion of cytidine 5′-monophosphate to cytidine 5′-triphosphate. In the whole cell biocatalysis, recombinant CMP kinase catalyzed the conversion of CMP to CDP, and endogenous acetate kinase of the E. coli was utilized for the ATP regeneration as well as for the conversion of CDP to CTP. A conversion yield of ca. 88 % CTP was obtained when starting with 20 mM CMP, 1 mM ATP, and 80 mM acetyl phosphate based on the initial CMP concentration. Endogenous pyruvate kinase and poly- phosphate kinase were inefficient in the process. The CTP production system was applied to the production of CMP-NeuAc by additionally introducing the CMP-NeuAc synthetase gene into the recombinant E. coli. Keywords: CMP-kinase, whole cell biocatalysis, ATP regeneration, cytidine 5′-monophosphate Introduction employing enzymes [4]. They applied various enzymatic 1) methods and chemical methods and concluded that the As glycosyltransferase-catalyzed synthetic techniques enzymatic method based on adenylate kinase/pyruvate are becoming recognized as powerful methods for the kinase provided the most convenient route to CTP. In the preparation of biologically important oligosaccharides, process, CTP was generated efficiently from an inexpen- the development of cost-efficient production methods for sive substrate, CMP. -
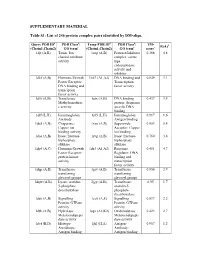
List of 246 Protein Complex Pairs Identified by DM-Align
SUPPLEMENTARY MATERIAL Table S1: List of 246 protein complex pairs identified by DM-align. Query PDB IDa PDB Classb: Temp PDB IDd PDB Classb: TM- R(Å)f (Chain1,Chain2) GO termc (Chain1,Chain2) GO termc scoree 1djt (A,B) Toxin: Ion 1aap (A,B) Protease/Inhibitor 0.368 4.8 channel inhibitor complex: serine activity type endopeptidase activity and inhibitor 1dkf (A,B) Hormone/Growth 1xb7 (A1,A2) DNA binding and 0.849 3.1 Factor Receptor: Transcription DNA binding and factor activity transciption factor activity 1dl5 (A,B) Transferase: 1utx (A,B) DNA binding 0.437 3.9 Methyltransferas protein: Sequence e activity specific DNA binding 1dlf (L,H) Immunoglobin: 1j05 (L,H) Immunoglobin: 0.917 1.6 Antibody Antigen binding 1do5 (A,B) Chaperone: 1xso (A,B) Superoxide 0.953 0.9 Copper ion Acceptor: Copper binding activity ion binding 1dos (A,B) lyase: fructose- 1rvg (A,B) lyase: fructose- 0.760 3.6 biphosphate biphosphate aldolase aldolase 1dp4 (A,C) Hormone/Growth 1dz3 (A1,A2) Response 0.481 4.7 Factor Receptor: Regulator: DNA protein kinase binding and activity transcription factor activity 1dqp (A,B) Transferase: 1grv (A,B) Transferase: 0.856 2.9 transferring transferring glycosyl groups glycosyl groups 1dqw (A,B) Lyase: orotidin- 2jgy (A,B) Transferase: 0.95 1.7 5-phosphate orotidin-5- decarboxylase phosphate decarboxylase 1ds6 (A,B) Signalling 1cc0 (A,E) Signalling 0.897 2.2 Protein: GTPase Protein: GTPase activity activity 1dth (A,B) Hydrolase: 1sqv (A2,K2) Oxidoredutase: 0.423 2.7 Metaloendopepti Metaloendopepti dase activity dase activity 1dvf -

Découverte D'une Nouvelle Famille De Protéine Kinases Bactériennes
Découverte d’une nouvelle famille de protéine kinases bactériennes : mécanismes de fonctionnement et rôle cellulaire de YdiB, un archétype chez Baccillus subtilis Hien-Anh Nguyen To cite this version: Hien-Anh Nguyen. Découverte d’une nouvelle famille de protéine kinases bactériennes : mécanismes de fonctionnement et rôle cellulaire de YdiB, un archétype chez Baccillus subtilis. Sciences agricoles. Université de Grenoble, 2012. Français. NNT : 2012GRENV017. tel-00721757 HAL Id: tel-00721757 https://tel.archives-ouvertes.fr/tel-00721757 Submitted on 30 Jul 2012 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE Pour obtenir le grade de DOCTEUR DE L’UNIVERSITÉ DE GRENOBLE Spécialité : Chimie-Biologie Arrêté ministériel : 7 août 2006 Présentée par Hien-Anh NGUYEN Thèse dirigée par le Dr. Jean-Michel JAULT préparée au sein de l’Institut de Biologie Structurale J.-P. Ebel, et du CEA de Grenoble dans l'École Doctorale Chimie et Sciences du Vivant Découverte d’une nouvelle famille de protéines kinases bactériennes : Mécanisme de fonctionnement et rôle cellulaire de YdiB, un représentant chez B. subtilis Thèse soutenue publiquement le 23 mai 2012 devant le jury composé de : Mme. Patricia DOUBLET Rapporteur Prof. -

Two-Dimensional Isobutyl Acetate Production Pathways to Improve Carbon Yield
ARTICLE Received 23 Apr 2015 | Accepted 13 May 2015 | Published 25 Jun 2015 DOI: 10.1038/ncomms8488 OPEN Two-dimensional isobutyl acetate production pathways to improve carbon yield Yohei Tashiro1, Shuchi H. Desai1,2 & Shota Atsumi1,2 For an economically competitive biological process, achieving high carbon yield of a target chemical is crucial. In biochemical production, pyruvate and acetyl-CoA are primary building blocks. When sugar is used as the sole biosynthetic substrate, acetyl-CoA is commonly generated by pyruvate decarboxylation. However, pyruvate decarboxylation during acetyl-CoA formation limits the theoretical maximum carbon yield (TMCY) by releasing carbon, and in some cases also leads to redox imbalance. To avoid these problems, we describe here the construction of a metabolic pathway that simultaneously utilizes glucose and acetate. Acetate is utilized to produce acetyl-CoA without carbon loss or redox imbalance. We demonstrate the utility of this approach for isobutyl acetate (IBA) production, wherein IBA production with glucose and acetate achieves a higher carbon yield than with either sole carbon source. These results highlight the potential for this multiple carbon source approach to improve the TMCY and balance redox in biosynthetic pathways. 1 Department of Chemistry, University of California, Davis, One Shields Avenue, Davis, California 95616, USA. 2 Microbiology Graduate Group, University of California, Davis, One Shields Avenue, Davis, California 95616, USA. Correspondence and requests for materials should be addressed to S.A. (email: [email protected]). NATURE COMMUNICATIONS | 6:7488 | DOI: 10.1038/ncomms8488 | www.nature.com/naturecommunications 1 & 2015 Macmillan Publishers Limited. All rights reserved. ARTICLE NATURE COMMUNICATIONS | DOI: 10.1038/ncomms8488 iochemical production from biomass, a renewable resource OH synthesized from CO2 and sunlight, may contribute to a 1 Bcarbon-neutral society . -

Acetate Activation in Methanosaeta Thermophila: Characterization of the Key Enzymes Pyrophosphatase and Acetyl-Coa Synthetase
Hindawi Publishing Corporation Archaea Volume 2012, Article ID 315153, 10 pages doi:10.1155/2012/315153 Research Article Acetate Activation in Methanosaeta thermophila: Characterization of the Key Enzymes Pyrophosphatase and Acetyl-CoA Synthetase Stefanie Berger, Cornelia Welte, and Uwe Deppenmeier Institute for Microbiology and Biotechnology, University of Bonn, Meckenheimer Allee 168, 53115 Bonn, Germany Correspondence should be addressed to Uwe Deppenmeier, [email protected] Received 16 May 2012; Accepted 30 June 2012 Academic Editor: Francesca Paradisi Copyright © 2012 Stefanie Berger et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The thermophilic methanogen Methanosaeta thermophila uses acetate as sole substrate for methanogenesis. It was proposed that the acetate activation reaction that is needed to feed acetate into the methanogenic pathway requires the hydrolysis of two ATP, whereas the acetate activation reaction in Methanosarcina sp. is known to require only one ATP. As these organisms live at the thermodynamic limit that sustains life, the acetate activation reaction in Mt. thermophila seems too costly and was thus reevaluated. It was found that of the putative acetate activation enzymes one gene encoding an AMP-forming acetyl-CoA synthetase was highly expressed. The corresponding enzyme was purified and characterized in detail. It catalyzed the ATP-dependent formation of acetyl- CoA, AMP, and pyrophosphate (PPi) and was only moderately inhibited by PPi. The breakdown of PPi was performed by a soluble pyrophosphatase. This enzyme was also purified and characterized. The pyrophosphatase hydrolyzed the major part of PPi (KM = 0.27 ± 0.05 mM) that was produced in the acetate activation reaction. -

Table S1. List of Oligonucleotide Primers Used
Table S1. List of oligonucleotide primers used. Cla4 LF-5' GTAGGATCCGCTCTGTCAAGCCTCCGACC M629Arev CCTCCCTCCATGTACTCcgcGATGACCCAgAGCTCGTTG M629Afwd CAACGAGCTcTGGGTCATCgcgGAGTACATGGAGGGAGG LF-3' GTAGGCCATCTAGGCCGCAATCTCGTCAAGTAAAGTCG RF-5' GTAGGCCTGAGTGGCCCGAGATTGCAACGTGTAACC RF-3' GTAGGATCCCGTACGCTGCGATCGCTTGC Ukc1 LF-5' GCAATATTATGTCTACTTTGAGCG M398Arev CCGCCGGGCAAgAAtTCcgcGAGAAGGTACAGATACGc M398Afwd gCGTATCTGTACCTTCTCgcgGAaTTcTTGCCCGGCGG LF-3' GAGGCCATCTAGGCCATTTACGATGGCAGACAAAGG RF-5' GTGGCCTGAGTGGCCATTGGTTTGGGCGAATGGC RF-3' GCAATATTCGTACGTCAACAGCGCG Nrc2 LF-5' GCAATATTTCGAAAAGGGTCGTTCC M454Grev GCCACCCATGCAGTAcTCgccGCAGAGGTAGAGGTAATC M454Gfwd GATTACCTCTACCTCTGCggcGAgTACTGCATGGGTGGC LF-3' GAGGCCATCTAGGCCGACGAGTGAAGCTTTCGAGCG RF-5' GAGGCCTGAGTGGCCTAAGCATCTTGGCTTCTGC RF-3' GCAATATTCGGTCAACGCTTTTCAGATACC Ipl1 LF-5' GTCAATATTCTACTTTGTGAAGACGCTGC M629Arev GCTCCCCACGACCAGCgAATTCGATagcGAGGAAGACTCGGCCCTCATC M629Afwd GATGAGGGCCGAGTCTTCCTCgctATCGAATTcGCTGGTCGTGGGGAGC LF-3' TGAGGCCATCTAGGCCGGTGCCTTAGATTCCGTATAGC RF-5' CATGGCCTGAGTGGCCGATTCTTCTTCTGTCATCGAC RF-3' GACAATATTGCTGACCTTGTCTACTTGG Ire1 LF-5' GCAATATTAAAGCACAACTCAACGC D1014Arev CCGTAGCCAAGCACCTCGgCCGAtATcGTGAGCGAAG D1014Afwd CTTCGCTCACgATaTCGGcCGAGGTGCTTGGCTACGG LF-3' GAGGCCATCTAGGCCAACTGGGCAAAGGAGATGGA RF-5' GAGGCCTGAGTGGCCGTGCGCCTGTGTATCTCTTTG RF-3' GCAATATTGGCCATCTGAGGGCTGAC Kin28 LF-5' GACAATATTCATCTTTCACCCTTCCAAAG L94Arev TGATGAGTGCTTCTAGATTGGTGTCggcGAAcTCgAGCACCAGGTTG L94Afwd CAACCTGGTGCTcGAgTTCgccGACACCAATCTAGAAGCACTCATCA LF-3' TGAGGCCATCTAGGCCCACAGAGATCCGCTTTAATGC RF-5' CATGGCCTGAGTGGCCAGGGCTAGTACGACCTCG -

Using MALDI-TOF MS Analysis for Rapid Discrimination of MRSA MLST
Using MALDI-TOF MS analysis for rapid discrimination of MRSA MLST types Jang-Jih Lu1,2,Tsui-Ping Liu1, Shih-Cheng Chang1,2 1Department of Laboratory Medicine, Chang Gung Memorial Hospital, Linkou, Taoyuan, Taiwan 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan Email: [email protected] 73 0:F8 MS Ra w 71 0:F9 MS Ra w 5000 6422.794 6000 Intens.[a.u.] Intens.[a.u.] Introduction 4000 Results ST5 ST5 4000 3000 2978.985 2000 6551.531 Staphylococcus aureus is one of the common causes of 3027.802 3054.785 2964.900 3175.943 2000 Table 1: MRSA and MSSA associated peaks by MALDI TOF 1000 community-associated infections and health care related 234 0:F1 MS Ra w 234 0:F1 MS Ra w 3005.919 MRSA(129 isolates) MSSA(77 isolates) 5000 6000 Intens.[a.u.] Intens.[a.u.] pathogen. It can usually cause wound infection, ST45 4000 ST45 6549.206 4000 3000 MS peaks (m/z) no. of isolate % of isolate no. of isolate % of isolate 6420.525 osteomyelitis, and even serious invasive infections, such as 2000 6610.628 2000 2978.373 3053.663 3174.805 2415 67 51.9 2 2.6 1000 sepsis and pneumonia. Epidemiology studies have shown 86 0:E4 MS Ra w 70 0:E5 MS Ra w 3058.565 5000 2431 65 50.4 0 0 3074.390 6000 Intens.[a.u.] Intens.[a.u.] that there are close relationship between the results of ST59 4000 6550.723 ST59 4000 3000 2879 51 39.5 2 2.6 6422.151 genotyping from multilocus sequence typing (MLST) and 2000 2000 characteristics of MRSA strains, including hospital- 6593 77 59.7 7 9.1 1000 200 0:E8 MS Ra w 38 0:E11 MS Ra w Appear of any above 103 79.8 11 14.3 2978.890 5000 6000 Intens.[a.u.] Intens.[a.u.] 6421.658 acquired, community-associated strain and even the 2965.568 ST239 4000 ST239 peaks 4000 3000 none of above peaks 6590.270 26 20.1 66 85.7 2937.113 3177.333 2916.384 antibiogram of the strain. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Saccharomyces Cerevisiae Aspartate Kinase Mechanism and Inhibition
In compliance with the Canadian Privacy Legislation some supporting forms may have been removed from this dissertation. While these forms may be included in the document page count, their removal does not represent any loss of content from the dissertation. Ph.D. Thesis - D. Bareich McMaster University - Department of Biochemistry FUNGAL ASPARTATE KINASE MECHANISM AND INHIBITION By DAVID C. BAREICH, B.Sc. A Thesis Submitted to the School of Graduate Studies in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy McMaster University © Copyright by David C. Bareich, June 2003 1 Ph.D. Thesis - D. Bareich McMaster University - Department of Biochemistry FUNGAL ASPARTATE KINASE MECHANISM AND INHIBITION Ph.D. Thesis - D. Bareich McMaster University - Department of Biochemistry DOCTOR OF PHILOSOPHY (2003) McMaster University (Biochemistry) Hamilton, Ontario TITLE: Saccharomyces cerevisiae aspartate kinase mechanism and inhibition AUTHOR: David Christopher Bareich B.Sc. (University of Waterloo) SUPERVISOR: Professor Gerard D. Wright NUMBER OF PAGES: xix, 181 11 Ph.D. Thesis - D. Bareich McMaster University - Department of Biochemistry ABSTRACT Aspartate kinase (AK) from Saccharomyces cerevisiae (AKsc) catalyzes the first step in the aspartate pathway responsible for biosynthesis of L-threonine, L-isoleucine, and L-methionine in fungi. Little was known about amino acids important for AKsc substrate binding and catalysis. Hypotheses about important amino acids were tested using site directed mutagenesis to substitute these amino acids with others having different properties. Steady state kinetic parameters and pH titrations of the variant enzymes showed AKsc-K18 and H292 to be important for binding and catalysis. Little was known about how the S. cerevisiae aspartate pathway kinases, AKsc and homoserine kinase (HSKsc), catalyze the transfer of the y-phosphate from adenosine triphosphate (ATP) to L-aspartate or L-homoserine, respectively. -

Supplementary Information
Supplementary information (a) (b) Figure S1. Resistant (a) and sensitive (b) gene scores plotted against subsystems involved in cell regulation. The small circles represent the individual hits and the large circles represent the mean of each subsystem. Each individual score signifies the mean of 12 trials – three biological and four technical. The p-value was calculated as a two-tailed t-test and significance was determined using the Benjamini-Hochberg procedure; false discovery rate was selected to be 0.1. Plots constructed using Pathway Tools, Omics Dashboard. Figure S2. Connectivity map displaying the predicted functional associations between the silver-resistant gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S3. Connectivity map displaying the predicted functional associations between the silver-sensitive gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S4. Metabolic overview of the pathways in Escherichia coli. The pathways involved in silver-resistance are coloured according to respective normalized score. Each individual score represents the mean of 12 trials – three biological and four technical. Amino acid – upward pointing triangle, carbohydrate – square, proteins – diamond, purines – vertical ellipse, cofactor – downward pointing triangle, tRNA – tee, and other – circle. -

Q 297 Suppl USE
The following supplement accompanies the article Atlantic salmon raised with diets low in long-chain polyunsaturated n-3 fatty acids in freshwater have a Mycoplasma dominated gut microbiota at sea Yang Jin, Inga Leena Angell, Simen Rød Sandve, Lars Gustav Snipen, Yngvar Olsen, Knut Rudi* *Corresponding author: [email protected] Aquaculture Environment Interactions 11: 31–39 (2019) Table S1. Composition of high- and low LC-PUFA diets. Stage Fresh water Sea water Feed type High LC-PUFA Low LC-PUFA Fish oil Initial fish weight (g) 0.2 0.4 1 5 15 30 50 0.2 0.4 1 5 15 30 50 80 200 Feed size (mm) 0.6 0.9 1.3 1.7 2.2 2.8 3.5 0.6 0.9 1.3 1.7 2.2 2.8 3.5 3.5 4.9 North Atlantic fishmeal (%) 41 40 40 40 40 30 30 41 40 40 40 40 30 30 35 25 Plant meals (%) 46 45 45 42 40 49 48 46 45 45 42 40 49 48 39 46 Additives (%) 3.3 3.2 3.2 3.5 3.3 3.4 3.9 3.3 3.2 3.2 3.5 3.3 3.4 3.9 2.6 3.3 North Atlantic fish oil (%) 9.9 12 12 15 16 17 18 0 0 0 0 0 1.2 1.2 23 26 Linseed oil (%) 0 0 0 0 0 0 0 6.8 8.1 8.1 9.7 11 10 11 0 0 Palm oil (%) 0 0 0 0 0 0 0 3.2 3.8 3.8 5.4 5.9 5.8 5.9 0 0 Protein (%) 56 55 55 51 49 47 47 56 55 55 51 49 47 47 44 41 Fat (%) 16 18 18 21 22 22 22 16 18 18 21 22 22 22 28 31 EPA+DHA (% diet) 2.2 2.4 2.4 2.9 3.1 3.1 3.1 0.7 0.7 0.7 0.7 0.7 0.7 0.7 4 4.2 Table S2.