Small Tropical Islands with Dense Human Population: Differences in Water Quality of Near-Shore Waters Are Associated with Distinct Bacterial Communities
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
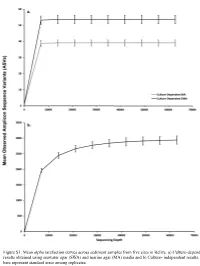
Figure S1. Mean Alpha Rarefaction Curves Across Sediment Samples from Five Sites in Belize
Figure S1. Mean alpha rarefaction curves across sediment samples from five sites in Belize. a) Culture-dependent results obtained using seawater agar (SWA) and marine agar (MA) media and b) Culture- independent results. Error bars represent standard error among replicates. a. 200 150 100 Faith’s PD Faith’s 50 0 Culturedependent MA Culturedependent SWA Cultureindependent Method b. 0.8 0.6 Pielou’s Evenness 0.4 Culturedependent MA Culturedependent SWA Cultureindependent Method Figure S2. Alpha diversity boxplots of marine sediment microbial communities from Carrie Bow Cay, Belize in culture-dependent and culture-independent samples determined using a) Faith’s Phylogenetic Diversity Index and b) Pielou’s Evenness. Culture-dependent methods include the use of marine agar medium (MA) and seawater agar medium (SWA. Data points are overlayed on the boxplot to show variation. a. 100% Proteobacteria Nitrospinae Bacteroidetes Fibrobacteres Planctomycetes Entotheonellaeota 90% Cyanobacteria Marinimicrobia (SAR406 clade) Firmicutes Deinococcus-Thermus Chloroflexi Margulisbacteria [A] Thaumarchaeota Unassigned 80% Acidobacteria Archaea UA Verrucomicrobia Acetothermia Actinobacteria Elusimicrobia 70% [A] Nanoarchaeaeota Tenericutes Kiritimatiellaeota TA06 Latescibacteria WS2 60% Spirochaetes Dependentiae Patescibacteria LCP-89 Gemmatimonadetes FCPU426 Omnitrophicaeota WPS-2 50% Fusobacteria CK-2C2-2 Bacteria UA Armatimonadetes [A] Euryarchaeota [A] Altiarchaeota Relative Percent 40% Calditrichaeota Poribacteria Lentisphaerae Cloacimonetes [A] Crenarchaeota -

Flavobacterial Response to Organic Pollution
Vol. 51: 31–43, 2008 AQUATIC MICROBIAL ECOLOGY Published April 24 doi: 10.3354/ame01174 Aquat Microb Ecol Flavobacterial response to organic pollution Andrew Bissett1, 2, 3,*, John P. Bowman2, Chris M. Burke1 1School of Aquaculture, Tasmanian Aquaculture and Fisheries Institute, University of Tasmania and Aquafin CRC, Launceston, Tasmania 7250, Australia 2School of Agricultural Science, University of Tasmania, Hobart, Tasmania 7001, Australia 3Present address: Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, 28359 Bremen, Germany ABSTRACT: Bacteria of the Cytophaga-Flavobacterium-Bacteroides (CFB) group (phylum Bac- teroidetes), in particular members of the class Flavobacteria, are among the most prominent hetero- trophic organisms in marine pelagic systems. They have also previously been found to be important in the initial biopolymer degradation of sedimentary organic matter. The Flavobacteria community was analysed in inshore, marine sediments subject to regular inputs of highly labile organic carbon in order to understand the importance of this group in carbon degradation. We used denaturing gra- dient gel electrophoresis and real-time PCR in a statistically robust manner, over 2 consecutive years, to demonstrate that the number of Flavobacteria in the sediment increased and community composi- tion shifted with organic loading. Further community shifts occurred after cessation of organic load- ing, and population numbers also decreased. Flavobacteria appear to be important in the initial responses of the sediment microbial community to organic loading, regardless of sediment type, but flavobacterial composition was not predictable. The highly dynamic nature and large diversity (func- tional redundancy) of the Flavobacteria in these sediments may contribute to this unpredictable response. KEY WORDS: Flavobacteria . -

Compile.Xlsx
Silva OTU GS1A % PS1B % Taxonomy_Silva_132 otu0001 0 0 2 0.05 Bacteria;Acidobacteria;Acidobacteria_un;Acidobacteria_un;Acidobacteria_un;Acidobacteria_un; otu0002 0 0 1 0.02 Bacteria;Acidobacteria;Acidobacteriia;Solibacterales;Solibacteraceae_(Subgroup_3);PAUC26f; otu0003 49 0.82 5 0.12 Bacteria;Acidobacteria;Aminicenantia;Aminicenantales;Aminicenantales_fa;Aminicenantales_ge; otu0004 1 0.02 7 0.17 Bacteria;Acidobacteria;AT-s3-28;AT-s3-28_or;AT-s3-28_fa;AT-s3-28_ge; otu0005 1 0.02 0 0 Bacteria;Acidobacteria;Blastocatellia_(Subgroup_4);Blastocatellales;Blastocatellaceae;Blastocatella; otu0006 0 0 2 0.05 Bacteria;Acidobacteria;Holophagae;Subgroup_7;Subgroup_7_fa;Subgroup_7_ge; otu0007 1 0.02 0 0 Bacteria;Acidobacteria;ODP1230B23.02;ODP1230B23.02_or;ODP1230B23.02_fa;ODP1230B23.02_ge; otu0008 1 0.02 15 0.36 Bacteria;Acidobacteria;Subgroup_17;Subgroup_17_or;Subgroup_17_fa;Subgroup_17_ge; otu0009 9 0.15 41 0.99 Bacteria;Acidobacteria;Subgroup_21;Subgroup_21_or;Subgroup_21_fa;Subgroup_21_ge; otu0010 5 0.08 50 1.21 Bacteria;Acidobacteria;Subgroup_22;Subgroup_22_or;Subgroup_22_fa;Subgroup_22_ge; otu0011 2 0.03 11 0.27 Bacteria;Acidobacteria;Subgroup_26;Subgroup_26_or;Subgroup_26_fa;Subgroup_26_ge; otu0012 0 0 1 0.02 Bacteria;Acidobacteria;Subgroup_5;Subgroup_5_or;Subgroup_5_fa;Subgroup_5_ge; otu0013 1 0.02 13 0.32 Bacteria;Acidobacteria;Subgroup_6;Subgroup_6_or;Subgroup_6_fa;Subgroup_6_ge; otu0014 0 0 1 0.02 Bacteria;Acidobacteria;Subgroup_6;Subgroup_6_un;Subgroup_6_un;Subgroup_6_un; otu0015 8 0.13 30 0.73 Bacteria;Acidobacteria;Subgroup_9;Subgroup_9_or;Subgroup_9_fa;Subgroup_9_ge; -

The Intestinal Bacterial Community and Functional Potential of Litopenaeus Vannamei in the Coastal Areas of China
microorganisms Article The Intestinal Bacterial Community and Functional Potential of Litopenaeus vannamei in the Coastal Areas of China Yimeng Cheng 1, Chaorong Ge 1,*, Wei Li 1 and Huaiying Yao 1,2,3 1 Research Center for Environmental Ecology and Engineering, School of Environmental Ecology and Biological Engineering, Wuhan Institute of Technology, Wuhan 430073, China; [email protected] (Y.C.); [email protected] (W.L.); [email protected] (H.Y.) 2 Zhejiang Key Laboratory of Urban Environmental Processes and Pollution Control, Ningbo Urban Environment Observation and Research Station, Chinese Academy of Sciences, Ningbo 315800, China 3 Key Laboratory of Urban Environment and Health, Institute of Urban Environment, Chinese Academy of Sciences, Xiamen 361021, China * Correspondence: [email protected] Abstract: Intestinal bacteria are crucial for the healthy aquaculture of Litopenaeus vannamei, and the coastal areas of China are important areas for concentrated L. vannamei cultivation. In this study, we evaluated different compositions and structures, key roles, and functional potentials of the intestinal bacterial community of L. vannamei shrimp collected in 12 Chinese coastal cities and investigated the correlation between the intestinal bacteria and functional potentials. The dominant bacteria in the shrimp intestines included Proteobacteria, Bacteroidetes, Tenericutes, Firmicutes, and Actinobacteria, and the main potential functions were metabolism, genetic information processing, and environmental information processing. Although the composition and structure of the intestinal bacterial community, potential pathogenic bacteria, and spoilage organisms varied from region to region, the functional potentials were homeostatic and significantly (p < 0.05) correlated with Citation: Cheng, Y.; Ge, C.; Li, W.; intestinal bacteria (at the family level) to different degrees. -

Contents Topic 1. Introduction to Microbiology. the Subject and Tasks
Contents Topic 1. Introduction to microbiology. The subject and tasks of microbiology. A short historical essay………………………………………………………………5 Topic 2. Systematics and nomenclature of microorganisms……………………. 10 Topic 3. General characteristics of prokaryotic cells. Gram’s method ………...45 Topic 4. Principles of health protection and safety rules in the microbiological laboratory. Design, equipment, and working regimen of a microbiological laboratory………………………………………………………………………….162 Topic 5. Physiology of bacteria, fungi, viruses, mycoplasmas, rickettsia……...185 TOPIC 1. INTRODUCTION TO MICROBIOLOGY. THE SUBJECT AND TASKS OF MICROBIOLOGY. A SHORT HISTORICAL ESSAY. Contents 1. Subject, tasks and achievements of modern microbiology. 2. The role of microorganisms in human life. 3. Differentiation of microbiology in the industry. 4. Communication of microbiology with other sciences. 5. Periods in the development of microbiology. 6. The contribution of domestic scientists in the development of microbiology. 7. The value of microbiology in the system of training veterinarians. 8. Methods of studying microorganisms. Microbiology is a science, which study most shallow living creatures - microorganisms. Before inventing of microscope humanity was in dark about their existence. But during the centuries people could make use of processes vital activity of microbes for its needs. They could prepare a koumiss, alcohol, wine, vinegar, bread, and other products. During many centuries the nature of fermentations remained incomprehensible. Microbiology learns morphology, physiology, genetics and microorganisms systematization, their ecology and the other life forms. Specific Classes of Microorganisms Algae Protozoa Fungi (yeasts and molds) Bacteria Rickettsiae Viruses Prions The Microorganisms are extraordinarily widely spread in nature. They literally ubiquitous forward us from birth to our death. Daily, hourly we eat up thousands and thousands of microbes together with air, water, food. -

Centro De Investigación En Alimentación Y Desarrollo, A.C
Centro de Investigación en Alimentación y Desarrollo, A.C. CARACTERIZACIÓN DE LA COMUNIDAD BACTERIANA DEL CORAL POCILLOPORA CAPITATA DE LOS PARCHES CORALINOS DE MANZANILLO, COLIMA. Por: Lic. María de los Angeles Milagros Laurel Sandoval TESIS APROBADA POR LA: UNIDAD MAZATLÁN EN ACUICULTURA Y MANEJO AMBIENTAL Como requisito parcial para obtener el grado de MAESTRÍA EN CIENCIAS Mazatlán, Sinaloa. Enero, 2014 ii DECLARACIÓN INSTITUCIONAL La información generada en esta tesis es propiedad intelectual del Centro de Investigación en Alimentación y Desarrollo, A.C. Se permiten y agradecen las citas breves del material contenido en esta tesis sin permiso especial del autor, siempre y cuando se dé crédito correspondiente. Para la reproducción parcial o total de la tesis con fines académicos, se deberá contar con la autorización escrita del director del Centro de Investigación en Alimentación y Desarrollo, A.C. (CIAD). La publicación en comunicaciones científicas o de divulgación popular de los datos contenidos en esta tesis, deberá dar los créditos al CIAD, previa autorización escrita del manuscrito en cuestión del director de tesis. Dr. Pablo Wong González Director General iii AGRADECIEMIENTOS Al Consejo Nacional de Ciencia y Tecnología (CONACyT) por el apoyo económico brindado durante mi maestría en el CIAD, así como también por la beca mixta asignada para la realización de la estancia en en la Universidad del Estado de San Diego (SDSU). Al Centro de Investigación en Alimentación y Desarrollo (CIAD), por permitirme la oportunidad de llevar a cabo mis estudios de maestría en su programa de posgrado. A mi directora de tesis la Dra. Sonia Araceli Soto Rodríguez por su confianza y apoyo en la realización de esta tesis, agradeciéndole sus aportaciones y tiempo para alcanzar esta meta. -

Bacterial Taxa Based on Greengenes Database GS1A PS1B ABY1 OD1
A1: Bacterial taxa based on GreenGenes database GS1A PS1B ABY1_OD1 0.1682 0.024 Bacteria;ABY1_OD1;ABY1_OD1_unclassified 1 0 Bacteria;ABY1_OD1;FW129;FW129_unclassified 4 0 Bacteria;ABY1_OD1;FW129;KNA6-NB12;KNA6-NB12_unclassified 5 0 Bacteria;ABY1_OD1;FW129;KNA6-NB29;KNA6-NB29_unclassified 0 1 Acidobacteria 0.7907 4.509 Bacteria;Acidobacteria;Acidobacteria_unclassified 4 31 Bacteria;Acidobacteria;Acidobacteria-5;Acidobacteria-5_unclassified 0 1 Bacteria;Acidobacteria;BPC015;BPC015_unclassified 8 30 Bacteria;Acidobacteria;BPC102;BPC102_unclassified 9 43 Bacteria;Acidobacteria;Chloracidobacteria;Ellin6075;Ellin6075_unclassified 1 0 Bacteria;Acidobacteria;iii1-15;Acidobacteria-6;RB40;RB40_unclassified 0 5 Bacteria;Acidobacteria;iii1-15;iii1-15_unclassified 1 8 Bacteria;Acidobacteria;iii1-15;Riz6I;Unclassified 0 1 Bacteria;Acidobacteria;iii1-8;Unclassified 0 2 Bacteria;Acidobacteria;OS-K;OS-K_unclassified 18 17 Bacteria;Acidobacteria;RB25;RB25_unclassified 6 47 Bacteria;Acidobacteria;Solibacteres;Solibacteres_unclassified 0 1 Actinobacteria 2.1198 6.642 Bacteria;Actinobacteria;Acidimicrobidae;Acidimicrobidae_unclassified 10 70 Bacteria;Actinobacteria;Acidimicrobidae;CL500-29;ML316M-15;ML316M-15_unclassified 0 3 Bacteria;Actinobacteria;Acidimicrobidae;EB1017_group;Acidimicrobidae_bacterium_Ellin7143;Unclassified 6 1 Bacteria;Actinobacteria;Acidimicrobidae;koll13;JTB31;BD2-10;BD2-10_unclassified 1 5 Bacteria;Actinobacteria;Acidimicrobidae;koll13;JTB31;Unclassified 16 37 Bacteria;Actinobacteria;Acidimicrobidae;koll13;koll13_unclassified 81 25 Bacteria;Actinobacteria;Acidimicrobidae;Microthrixineae;Microthrixineae_unclassified -

The Effect of Surface Colour on the Formation of Marine Micro- and Macro-Fouling Communities 5 Sergey Dobretsov1*, Raeid M
To Biofouling journal The effect of surface colour on the formation of marine micro- and macro-fouling communities 5 Sergey Dobretsov1*, Raeid M. M. Abed2 and Christian R. Voolstra3 1 Department of Marine Science and Fisheries, College of Agricultural and Marine Sciences, Sultan Qaboos University, Muscat, Oman 2 Department of Biology, College of Science, Sultan Qaboos University, Muscat, 10 Oman 3 Red Sea Research Center, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia * Corresponding author 15 Dr. Sergey Dobretsov Department of Marine Science and Fisheries, College of Agricultural and Marine Sciences, Sultan Qaboos University, PO Box 34 Al Koud 123, Sultanate Oman Tel.: (968) 24143657 Fax: (968) 24413418 20 e-mail: [email protected] [email protected] Running title: The effect of colour on biofouling communities 1 Abstract 25 In this study, the effect of substrate background colour on formation of micro- and macro-fouling communities was investigated. Acrylic tiles were painted with black and white colours and were covered with transparent sheets in order to ensure similar surface properties. All substrata were exposed to biofouling at 1 m depth for 40 days in the Marina Bandar al Rowdha (Muscat, Sea of Oman). Studies were conducted in 30 2010 over a time course of 5, 10, and 20 days, and in 2012 samples were collected at 7, 14, and 21 days. Bacterial densities on black and white substrata were similar except at day 10, when black substrata had the higher abundance than white ones. Pyrosequencing via 454 of 16S rRNA genes of bacteria from white and black substrata revealed that Alphaproteobacteria and Firmicutes were the dominant 35 groups. -

The Microbiome of the Egyptian Red Sea Proper and Gulf of Aqaba
American University in Cairo AUC Knowledge Fountain Theses and Dissertations 2-1-2016 The microbiome of The Egyptian Red Sea proper and Gulf of Aqaba Ghada Alaa El-Din Mustafa Follow this and additional works at: https://fount.aucegypt.edu/etds Recommended Citation APA Citation Mustafa, G. (2016).The microbiome of The Egyptian Red Sea proper and Gulf of Aqaba [Master’s thesis, the American University in Cairo]. AUC Knowledge Fountain. https://fount.aucegypt.edu/etds/33 MLA Citation Mustafa, Ghada Alaa El-Din. The microbiome of The Egyptian Red Sea proper and Gulf of Aqaba. 2016. American University in Cairo, Master's thesis. AUC Knowledge Fountain. https://fount.aucegypt.edu/etds/33 This Dissertation is brought to you for free and open access by AUC Knowledge Fountain. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of AUC Knowledge Fountain. For more information, please contact [email protected]. The American University in Cairo School of Sciences and Engineering THE MICROBIOME OF THE EGYPTIAN RED SEA PROPER AND GULF OF AQABA A Thesis Submitted to The Applied Sciences Graduate Program in partial fulfillment of the requirements for the degree of Doctorate in Applied Sciences (Biotechnology) By Ghada Alaa El-Din Kamal Mustafa Masters of Science- American University in Cairo Bachelor of Science - Ain Shams University Under the supervision of Professor Rania Siam Chair of the Biology Department Fall 2015 I The American University in Cairo The Microbiome of the Egyptian Red Sea Proper and Gulf of Aqaba A Thesis Submitted by Ghada Alaa El-Din Kamal Mustafa To the Biotechnology Graduate Program, Fall 2015 in partial fulfillment of the requirements for the degree of Doctorate of Applied Sciences in Biotechnology has been approved by Dr. -

A 1926 Supplement
Supplement to Trevathan-Tackett et al. (2020) – Aquat Microb Ecol 84:59-74 – https://doi.org/10.3354/ame01926 Table S1: Operational taxonomic units (OTUs) associated with seagrass or microalgae chloroplasts, and the BLAST results. Top BLAST Result: Alternative BLAST result: Greengenes Sequence Proportion of Accession Number; Proportion Identity; Accession Number; Proportion Identity; OTU ID result counts sequences Description Description KX808295.1; 1; Zostera noltei 18S ribosomal RNA (rrn18) gene, complete sequence; KX808392.1; 0.9951; Zostera marina voucher Otu40 * 867 0.001 mitochondrial C2544 mitochondrion, complete genome KX808295.1; 0.9378; Zostera noltei 18S ribosomal RNA (rrn18) gene, complete sequence; KX808392.1; 0.933; Zostera marina voucher Otu16 * 2687 0.002 mitochondrial C2544 mitochondrion, complete genome HM485224.1; 1; Uncultured microorganism clone Cyanobacteria PK_Seq406_EDW_339 small subunit ribosomal Otu11 Chloroplast 3628 0.002 RNA gene, partial sequence MF370229.1; 0.9631; Zostera marina chloroplast, Otu92 Chloroplast 837 0.001 complete genome KT973350.1; 1; Uncultured prokaryote clone KP792487.1; 0.9975; Bacillariophyta sp. 867-32 2986479 16S ribosomal RNA gene, partial small subunit ribosomal RNA gene, partial Otu190 Bacillariophyta 2177 0.001 sequence sequence; plastid JN232305.1; 0.9975; Uncultured diatom clone EU542369.1; 0.9951; Uncultured phototrophic 1LM8-18 16S ribosomal RNA gene, partial eukaryote clone SAV07H04 16S ribosomal RNA Otu15 Bacillariophyta 3291 0.002 sequence; plastid gene, partial sequence; plastid LC006286.1; 1; Uncultured cyanobacterium gene KP792462.1; 0.9975; Planoglabratella for 16S ribosomal RNA, partial sequence, clone: opercularis isolate GO_m14-08 small subunit Otu21 Bacillariophyta 8277 0.005 BZm-K31 ribosomal RNA gene, partial sequence; plastid EU376247.1; 1; Uncultured bacterium clone EU580477.1; 0.9975; Cymatopleura solea clone sls1807 16S ribosomal RNA gene, partial D14 66 16S ribosomal RNA gene, partial Otu438 Bacillariophyta 9118 0.006 sequence sequence; chloroplast FJ002196.1; 0.9926; Melosira cf. -

Host Species and Body Site Explain the Variation in the Microbiota Associated to Wild 2 Sympatric Mediterranean Teleost Fishes
Manuscript_2nd_rev Click here to access/download;Manuscript;teleostean_bacteria_rev2_def.do Click here to view linked References 1 Host species and body site explain the variation in the microbiota associated to wild 2 sympatric Mediterranean teleost fishes 3 4 M. Ruiz-Rodríguez1, M. Scheifler1, S. Sanchez-Brosseau1, E. Magnanou1, N. West2, M. 5 Suzuki2, S. Duperron3, Y. Desdevises1 6 1Sorbonne Université, CNRS, Biologie Intégrative des Organismes Marins, BIOM, 7 Observatoire Océanologique, F-66650 Banyuls/Mer, France. 8 2Sorbonne Université, CNRS, Observatoire Océanologique de Banyuls, FR3724, F- 9 66650 Banyuls/Mer, France. 10 3Muséum National d’Histoire Naturelle, CNRS, Molécules de Communication et 11 Adaptation des Micro-organismes, MCAM, Muséum National d’Histoire Naturelle, F- 12 75000 Paris, France. 13 14 Corresponding author: 15 M. Ruiz-Rodríguez. ORCID 0000-0002-4202-5180 16 E-mail address: [email protected] 17 Tel: +33 66 22 60 351 1 18 ABSTRACT 19 Microorganisms are an important component shaping the evolution of hosts and as such, 20 the study of bacterial communities with molecular techniques is shedding light on the 21 complexity of symbioses between bacteria and vertebrates. Teleost fish are a 22 heterogeneous group that live in a wide variety of habitats, and thus a good model group 23 to investigate symbiotic interactions and their influence on host biology and ecology. 24 Here we describe the microbiota of thirteen teleostean species sharing the same 25 environment in the Mediterranean Sea, and compare bacterial communities among 26 different species and body sites (external mucus, skin, gills and intestine). Our results 27 show that Proteobacteria is the dominant phylum present in fish and water. -

A Multilevel Assessment of Sediment Bioremediation with Bioturbating Macrofauna
A multilevel assessment of sediment bioremediation with bioturbating macrofauna Sebastian Vadillo Gonzalez A thesis submitted in fulfillment of the requirement for the degree of Doctor of Philosophy School of Biological, Earth and Environmental Sciences Faculty of Science Supervisors: Prof. Emma Johnston, Dr. Katherine Dafforn and Assoc Prof. Paul E. Gribben JANUARY 17, 2020 Thesis/Dissertation Sheet Surname/Family Name : Vadillo Gonzalez Given Name/s : Sebastian Abbreviation for degree as give in the : PhD University calendar Faculty : Science School : Biological, Earth and Environmental Sciences A multilevel approach on the bioremediation of eutrophic sediments using Thesis Title : benthic macrofauna Abstract 350 words maximum: (PLEASE TYPE) Estuaries and coastal intertidal environments supply important ecosystem services and resources. Within these systems, sediments play a crucial role in processing organic inputs and providing nutrients for local food webs. Benthic macrofauna and sediment microbial communities participate actively in these processes and can interact to influence overall ecosystem function. Through different bioturbation processes, macrofauna can affect biogeochemical cycling in sediments and influence microbial communities. However, anthropogenic derived eutrophication has increased globally in recent years and has been found to negatively impact these systems. Excess inputs of anthropogenic contaminants can disrupt key sediment processes involving macrofauna and microbial communities. Current options for sediment