George Charimba
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Eelgrass Sediment Microbiome As a Nitrous Oxide Sink in Brackish Lake Akkeshi, Japan
Microbes Environ. Vol. 34, No. 1, 13-22, 2019 https://www.jstage.jst.go.jp/browse/jsme2 doi:10.1264/jsme2.ME18103 Eelgrass Sediment Microbiome as a Nitrous Oxide Sink in Brackish Lake Akkeshi, Japan TATSUNORI NAKAGAWA1*, YUKI TSUCHIYA1, SHINGO UEDA1, MANABU FUKUI2, and REIJI TAKAHASHI1 1College of Bioresource Sciences, Nihon University, 1866 Kameino, Fujisawa, 252–0880, Japan; and 2Institute of Low Temperature Science, Hokkaido University, Kita-19, Nishi-8, Kita-ku, Sapporo, 060–0819, Japan (Received July 16, 2018—Accepted October 22, 2018—Published online December 1, 2018) Nitrous oxide (N2O) is a powerful greenhouse gas; however, limited information is currently available on the microbiomes involved in its sink and source in seagrass meadow sediments. Using laboratory incubations, a quantitative PCR (qPCR) analysis of N2O reductase (nosZ) and ammonia monooxygenase subunit A (amoA) genes, and a metagenome analysis based on the nosZ gene, we investigated the abundance of N2O-reducing microorganisms and ammonia-oxidizing prokaryotes as well as the community compositions of N2O-reducing microorganisms in in situ and cultivated sediments in the non-eelgrass and eelgrass zones of Lake Akkeshi, Japan. Laboratory incubations showed that N2O was reduced by eelgrass sediments and emitted by non-eelgrass sediments. qPCR analyses revealed that the abundance of nosZ gene clade II in both sediments before and after the incubation as higher in the eelgrass zone than in the non-eelgrass zone. In contrast, the abundance of ammonia-oxidizing archaeal amoA genes increased after incubations in the non-eelgrass zone only. Metagenome analyses of nosZ genes revealed that the lineages Dechloromonas-Magnetospirillum-Thiocapsa and Bacteroidetes (Flavobacteriia) within nosZ gene clade II were the main populations in the N2O-reducing microbiome in the in situ sediments of eelgrass zones. -

Polyphasic Study of Chryseobacterium Strains Isolated from Diseased Aquatic Animals Jean Francois Bernardet, M
Polyphasic study of Chryseobacterium strains isolated from diseased aquatic animals Jean Francois Bernardet, M. Vancanneyt, O. Matte-Tailliez, L. Grisez, L. Grisez, Patrick Tailliez, Chantal Bizet, M. Nowakowski, Brigitte Kerouault, J. Swings To cite this version: Jean Francois Bernardet, M. Vancanneyt, O. Matte-Tailliez, L. Grisez, L. Grisez, et al.. Polyphasic study of Chryseobacterium strains isolated from diseased aquatic animals. Systematic and Applied Microbiology, Elsevier, 2005, 28 (7), pp.640-660. 10.1016/j.syapm.2005.03.016. hal-02681942 HAL Id: hal-02681942 https://hal.inrae.fr/hal-02681942 Submitted on 1 Jun 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. ARTICLE IN PRESS Systematic and Applied Microbiology 28 (2005) 640–660 www.elsevier.de/syapm Polyphasic study of Chryseobacterium strains isolated from diseased aquatic animals J.-F. Bernardeta,Ã, M. Vancanneytb, O. Matte-Taillieza, L. Grisezc,1, P. Tailliezd, C. Bizete, M. Nowakowskie, B. Kerouaulta, J. Swingsb aInstitut National de la Recherche Agronomique, Unite´ de Virologie -

Chryseobacterium Gleum Urinary Tract Infection
Genes Review 2015 Vol.1, No.1, pp.1-5 DOI: 10.18488/journal.103/2015.1.1/103.1.1.5 © 2015 Asian Medical Journals. All Rights Reserved. CHRYSEOBACTERIUM GLEUM URINARY TRACT INFECTION † Ramya. T.G1 --- Sabitha Baby2 --- Pravin Das3 --- Geetha.R.K4 1,2,4Department of Microbiology, Karuna Medical College, Vilayodi, Chittur, Palakkad, India 3Department of Medicine, Karuna Medical College, Vilayodi, Chittur, Palakkad, India ABSTRACT Introduction: Chryseobacterium gleum is an uncommon pathogen in humans. It is a gram negative, nonfermenting bacterium distributed widely in soil and water. We present a case of urinary tract infection caused by Chryseobacterium gleum in a patient with right lower ureteric calculi. Case presentation: This case describes a 62- year-old male admitted for ureteric calculi to the Department of Urology in a tertiary care hospital in Kerala. A strain of Chryseobacterium gleum was isolated and confirmed by MALDI-TOF MS .The bacterium was sensitive to Piperacillin-Tazobactum (100/10µg ), Cefotaxime(30µg),Ceftazidime(30 µg ) and Ofloxacin(30 µg). It was resistant to Nitrofurantoin (300µg),Tobramycin(10µg),Gentamicin(30µg),Nalidixic acid(30µg) and Amikacin(30µg). Conclusion: Chryseobacterium gleum should be considered as a potential opportunistic and emerging pathogen. Resistance to a wide range of antibiotics such as aminoglycosides, penicillin, cephalosporins has been documented. In depth studies on Epidemiological, virulence and pathogenicity factors needs to be done for better diagnosis and management. Keywords: Chryseobacterium gleum, Calculi, Flexirubin pigment, MALDI-ToF MS, Non-fermenter, UTI. Contribution/ Originality This study documents the first case of Chryseobacterium gleum associated UTI in South India. 1. INTRODUCTION Chryseobacterium species are found ubiquitously in nature. -

The Prevalence of Foodborne Pathogenic Bacteria on Cutting Boards and Their Ecological Correlation with Background Biota
AIMS Microbiology, 2(2): 138-151. DOI: 10.3934/microbiol.2016.2.138 Received: 23 April 2016 Accepted: 19 May 2016 Published: 22 May 2016 http://www.aimspress.com/journal/microbiology Research article The prevalence of foodborne pathogenic bacteria on cutting boards and their ecological correlation with background biota Noor-Azira Abdul-Mutalib 1,2,3, Syafinaz Amin Nordin 2, Malina Osman 2, Ahmad Muhaimin Roslan 4, Natsumi Ishida 5, Kenji Sakai 5, Yukihiro Tashiro 5, Kosuke Tashiro 6, Toshinari Maeda 1, and Yoshihito Shirai 1,* 1 Department of Biological Functions and Engineering, Graduate School of Life Science and Systems Engineering, Kyushu Institute of Technology, 2-4 Hibikino, Wakamatsu-ku, Kitakyushu, Fukuoka 808-0196, Japan 2 Department of Medical Microbiology and Parasitology, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia 3 Department of Food Service and Management, Faculty of Food Science and Technology, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia 4 Department of Bioprocess Technology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia 5 Laboratory of Soil Microbiology, Faculty of Agriculture, Graduate School, Kyushu University, 6- 10-1 Hakozaki, Higashi-ku, Fukuoka 812-8581, Japan 6 Laboratory of Molecular Gene Technique, Faculty of Agriculture, Graduate School, Kyushu University, 6-10-1 Hakozaki, Higashi-ku, Fukuoka 812-8581, Japan * Correspondence: E-mail: [email protected]; Tel.: +6012-9196951; Fax: +603-89471182. Abstract: This study implemented the pyrosequencing technique and real-time quantitative PCR to determine the prevalence of foodborne pathogenic bacteria (FPB) and as well as the ecological correlations of background biota and FPB present on restaurant cutting boards (CBs) collected in Seri Kembangan, Malaysia. -

High Quality Permanent Draft Genome Sequence of Chryseobacterium Bovis DSM 19482T, Isolated from Raw Cow Milk
Lawrence Berkeley National Laboratory Recent Work Title High quality permanent draft genome sequence of Chryseobacterium bovis DSM 19482T, isolated from raw cow milk. Permalink https://escholarship.org/uc/item/4b48v7v8 Journal Standards in genomic sciences, 12(1) ISSN 1944-3277 Authors Laviad-Shitrit, Sivan Göker, Markus Huntemann, Marcel et al. Publication Date 2017 DOI 10.1186/s40793-017-0242-6 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Laviad-Shitrit et al. Standards in Genomic Sciences (2017) 12:31 DOI 10.1186/s40793-017-0242-6 SHORT GENOME REPORT Open Access High quality permanent draft genome sequence of Chryseobacterium bovis DSM 19482T, isolated from raw cow milk Sivan Laviad-Shitrit1, Markus Göker2, Marcel Huntemann3, Alicia Clum3, Manoj Pillay3, Krishnaveni Palaniappan3, Neha Varghese3, Natalia Mikhailova3, Dimitrios Stamatis3, T. B. K. Reddy3, Chris Daum3, Nicole Shapiro3, Victor Markowitz3, Natalia Ivanova3, Tanja Woyke3, Hans-Peter Klenk4, Nikos C. Kyrpides3 and Malka Halpern1,5* Abstract Chryseobacterium bovis DSM 19482T (Hantsis-Zacharov et al., Int J Syst Evol Microbiol 58:1024-1028, 2008) is a Gram-negative, rod shaped, non-motile, facultative anaerobe, chemoorganotroph bacterium. C. bovis is a member of the Flavobacteriaceae, a family within the phylum Bacteroidetes. It was isolated when psychrotolerant bacterial communities in raw milk and their proteolytic and lipolytic traits were studied. Here we describe the features of this organism, together with the draft genome sequence and annotation. The DNA G + C content is 38.19%. The chromosome length is 3,346,045 bp. It encodes 3236 proteins and 105 RNA genes. The C. bovis genome is part of the Genomic Encyclopedia of Type Strains, Phase I: the one thousand microbial genomes study. -
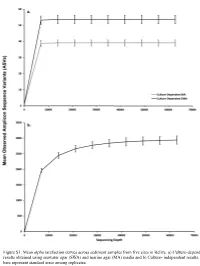
Figure S1. Mean Alpha Rarefaction Curves Across Sediment Samples from Five Sites in Belize
Figure S1. Mean alpha rarefaction curves across sediment samples from five sites in Belize. a) Culture-dependent results obtained using seawater agar (SWA) and marine agar (MA) media and b) Culture- independent results. Error bars represent standard error among replicates. a. 200 150 100 Faith’s PD Faith’s 50 0 Culturedependent MA Culturedependent SWA Cultureindependent Method b. 0.8 0.6 Pielou’s Evenness 0.4 Culturedependent MA Culturedependent SWA Cultureindependent Method Figure S2. Alpha diversity boxplots of marine sediment microbial communities from Carrie Bow Cay, Belize in culture-dependent and culture-independent samples determined using a) Faith’s Phylogenetic Diversity Index and b) Pielou’s Evenness. Culture-dependent methods include the use of marine agar medium (MA) and seawater agar medium (SWA. Data points are overlayed on the boxplot to show variation. a. 100% Proteobacteria Nitrospinae Bacteroidetes Fibrobacteres Planctomycetes Entotheonellaeota 90% Cyanobacteria Marinimicrobia (SAR406 clade) Firmicutes Deinococcus-Thermus Chloroflexi Margulisbacteria [A] Thaumarchaeota Unassigned 80% Acidobacteria Archaea UA Verrucomicrobia Acetothermia Actinobacteria Elusimicrobia 70% [A] Nanoarchaeaeota Tenericutes Kiritimatiellaeota TA06 Latescibacteria WS2 60% Spirochaetes Dependentiae Patescibacteria LCP-89 Gemmatimonadetes FCPU426 Omnitrophicaeota WPS-2 50% Fusobacteria CK-2C2-2 Bacteria UA Armatimonadetes [A] Euryarchaeota [A] Altiarchaeota Relative Percent 40% Calditrichaeota Poribacteria Lentisphaerae Cloacimonetes [A] Crenarchaeota -

Comparative Proteomic Profiling of Newly Acquired, Virulent And
www.nature.com/scientificreports OPEN Comparative proteomic profling of newly acquired, virulent and attenuated Neoparamoeba perurans proteins associated with amoebic gill disease Kerrie Ní Dhufaigh1*, Eugene Dillon2, Natasha Botwright3, Anita Talbot1, Ian O’Connor1, Eugene MacCarthy1 & Orla Slattery4 The causative agent of amoebic gill disease, Neoparamoeba perurans is reported to lose virulence during prolonged in vitro maintenance. In this study, the impact of prolonged culture on N. perurans virulence and its proteome was investigated. Two isolates, attenuated and virulent, had their virulence assessed in an experimental trial using Atlantic salmon smolts and their bacterial community composition was evaluated by 16S rRNA Illumina MiSeq sequencing. Soluble proteins were isolated from three isolates: a newly acquired, virulent and attenuated N. perurans culture. Proteins were analysed using two-dimensional electrophoresis coupled with liquid chromatography tandem mass spectrometry (LC–MS/MS). The challenge trial using naïve smolts confrmed a loss in virulence in the attenuated N. perurans culture. A greater diversity of bacterial communities was found in the microbiome of the virulent isolate in contrast to a reduction in microbial community richness in the attenuated microbiome. A collated proteome database of N. perurans, Amoebozoa and four bacterial genera resulted in 24 proteins diferentially expressed between the three cultures. The present LC–MS/ MS results indicate protein synthesis, oxidative stress and immunomodulation are upregulated in a newly acquired N. perurans culture and future studies may exploit these protein identifcations for therapeutic purposes in infected farmed fsh. Neoparamoeba perurans is an ectoparasitic protozoan responsible for the hyperplastic gill infection of marine cultured fnfsh referred to as amoebic gill disease (AGD)1. -

DNA Variation and Symbiotic Associations in Phenotypically Diverse Sea Urchin Strongylocentrotus Intermedius
DNA variation and symbiotic associations in phenotypically diverse sea urchin Strongylocentrotus intermedius Evgeniy S. Balakirev*†‡, Vladimir A. Pavlyuchkov§, and Francisco J. Ayala*‡ *Department of Ecology and Evolutionary Biology, University of California, Irvine, CA 92697-2525; †Institute of Marine Biology, Vladivostok 690041, Russia; and §Pacific Research Fisheries Centre (TINRO-Centre), Vladivostok, 690600 Russia Contributed by Francisco J. Ayala, August 20, 2008 (sent for review May 9, 2008) Strongylocentrotus intermedius (A. Agassiz, 1863) is an economically spines of the U form are relatively short; the length, as a rule, does important sea urchin inhabiting the northwest Pacific region of Asia. not exceed one third of the radius of the testa. The spines of the G The northern Primorye (Sea of Japan) populations of S. intermedius form are longer, reaching and frequently exceeding two thirds of the consist of two sympatric morphological forms, ‘‘usual’’ (U) and ‘‘gray’’ testa radius. The testa is significantly thicker in the U form than in (G). The two forms are significantly different in morphology and the G form. The morphological differences between the U and G preferred bathymetric distribution, the G form prevailing in deeper- forms of S. intermedius are stable and easily recognizable (Fig. 1), water settlements. We have analyzed the genetic composition of the and they are systematically reported for the northern Primorye S. intermedius forms using the nucleotide sequences of the mitochon- coast region (V.A.P., unpublished data). drial gene encoding the cytochrome c oxidase subunit I and the Little is known about the population genetics of S. intermedius; nuclear gene encoding bindin to evaluate the possibility of cryptic the available data are limited to allozyme polymorphisms (4–6). -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

08 Criobiology Rosario.P65
BIOCELL ISSN 0327 - 9545 2009, 33(3): A225-A241 PRINTED IN ARGENTINA Centro Binacional de Criobiología Clínica y Aplicada UNESCO Chair in Criobiology Facultad de Ciencias Bioquímicas y Farmacéuticas Universidad Nacional de Rosario Suipacha 570 (S2002LRK) Rosario Argentina Lectures and Abstracts from the 3er WORKSHOP IN CRYOBIOLOGY APPLIED TO MEDICAL SCIENCES May 5-7, 2009 Rosario, ARGENTINA The lectures and abstracts from 3er Workshop in Cryobiology of Medical Sciences have been revised and evaluated by a scientific committee A226 BIOCELL 33(3), 2009 Board of the 3er Workshop in Cryobiology Applied to Medical Sciences President Prof. Dr. Edgardo E. Guibert Vicepresident Prof. Dr. Joaquín V. Rodriguez Secretary Dra. Alejandra B. Quintana Sub-Secretary Dra. María Eugenia Mamprin Tresurer Dra. María Gabriela Mediavilla Collaborators Lic. María Dolores Pizarro Dra. Valeria Sigot Dra. Cecilia Mansilla Scientific Committee Dr. Blas Melissari (Uruguay) Dra. Alejandra Quintana (Argentina) Dra. María Eugenia Mamprin (Argentina) Dra. María Gabriela Mediavilla (Argentina) Dra. Valeria Sigot (Argentina) Governmental and Private sponsors ✓ Agencia Nacional de Promoción Científica y Tecnológica ✓ Secretaría de Ciencia, Tecnología e Innovación Productiva - SECYT ✓ Universidad Nacional de Rosario ✓ Gobierno de la Provincia de Santa Fe, Sub-Secretaría de Ciencia y Técnica ✓ UNESCO Chair in Cryobiology ✓ Cámara de Comercio Italiana de Rosario ✓ Consulado de Italia ✓ Centro Studi Fegato, Trieste, Italy ✓ AGA ✓ Claudio Lamelza S.R.L. ✓ Titania Viajes y Turismo -

| Hao Watatu Timur Alma Mult
|HAO WATATU US010058101B2TIMUR ALMA MULT (12 ) United States Patent ( 10 ) Patent No. : US 10 ,058 , 101 B2 von Maltzahn et al. (45 ) Date of Patent: * Aug. 28 , 2018 ( 54 ) METHODS OF USE OF SEED -ORIGIN ( 52 ) U . S . CI. ENDOPHYTE POPULATIONS CPC . .. .. .. AOIN 63 /00 (2013 .01 ) ; A01C 1 / 06 ( 2013 .01 ) ; A01N 63 / 02 ( 2013 .01 ) ; GOIN (71 ) Applicant: Indigo Ag , Inc. , Boston , MA (US ) 33/ 5097 ( 2013 . 01 ) ; A01C 1 / 60 ( 2013. 01 ) ( 72 ) Inventors : Geoffrey von Maltzahn , Boston , MA (58 ) Field of Classification Search (US ) ; Richard Bailey Flavell , None Thousand Oaks , CA (US ) ; Gerardo V . See application file for complete search history . Toledo , Belmont, MA (US ) ; Slavica Djonovic , Malden , MA (US ) ; Luis Miguel Marquez , Belmont, MA ( US) ; (56 ) References Cited David Morris Johnston , Cambridge , MA (US ) ; Yves Alain Millet, U . S . PATENT DOCUMENTS Newtonville , MA (US ) ; Jeffrey Lyford , 4 , 940 , 834 A 7 / 1990 Hurley et al . Hollis , NH (US ) ; Alexander Naydich , 5 ,041 , 290 A 8 / 1991 Gindrat et al. Cambridge , MA (US ) ; Craig Sadowski, 5 , 113 ,619 A 5 / 1992 Leps et al . 5 , 229 , 291 A * 7 / 1993 Nielsen . .. .. .. .. C12N 15 /00 Somerville , MA (US ) 435 / 244 5 , 292, 507 A 3 / 1994 Charley ( 73 ) Assignee : Indigo Agriculture, Inc. , Boston , MA 5 ,415 ,672 A 5 / 1995 Fahey et al . (US ) 5 ,730 , 973 A 3 / 1998 Morales et al. 5 , 994 , 117 A 11/ 1999 Bacon et al . 6 ,072 , 107 A 6 / 2000 Latch et al. ( * ) Notice : Subject to any disclaimer, the term of this 6 , 077 , 505 A 6 / 2000 Parke et al. -

Flavobacterial Response to Organic Pollution
Vol. 51: 31–43, 2008 AQUATIC MICROBIAL ECOLOGY Published April 24 doi: 10.3354/ame01174 Aquat Microb Ecol Flavobacterial response to organic pollution Andrew Bissett1, 2, 3,*, John P. Bowman2, Chris M. Burke1 1School of Aquaculture, Tasmanian Aquaculture and Fisheries Institute, University of Tasmania and Aquafin CRC, Launceston, Tasmania 7250, Australia 2School of Agricultural Science, University of Tasmania, Hobart, Tasmania 7001, Australia 3Present address: Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, 28359 Bremen, Germany ABSTRACT: Bacteria of the Cytophaga-Flavobacterium-Bacteroides (CFB) group (phylum Bac- teroidetes), in particular members of the class Flavobacteria, are among the most prominent hetero- trophic organisms in marine pelagic systems. They have also previously been found to be important in the initial biopolymer degradation of sedimentary organic matter. The Flavobacteria community was analysed in inshore, marine sediments subject to regular inputs of highly labile organic carbon in order to understand the importance of this group in carbon degradation. We used denaturing gra- dient gel electrophoresis and real-time PCR in a statistically robust manner, over 2 consecutive years, to demonstrate that the number of Flavobacteria in the sediment increased and community composi- tion shifted with organic loading. Further community shifts occurred after cessation of organic load- ing, and population numbers also decreased. Flavobacteria appear to be important in the initial responses of the sediment microbial community to organic loading, regardless of sediment type, but flavobacterial composition was not predictable. The highly dynamic nature and large diversity (func- tional redundancy) of the Flavobacteria in these sediments may contribute to this unpredictable response. KEY WORDS: Flavobacteria .