Flavobacterial Response to Organic Pollution
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Eelgrass Sediment Microbiome As a Nitrous Oxide Sink in Brackish Lake Akkeshi, Japan
Microbes Environ. Vol. 34, No. 1, 13-22, 2019 https://www.jstage.jst.go.jp/browse/jsme2 doi:10.1264/jsme2.ME18103 Eelgrass Sediment Microbiome as a Nitrous Oxide Sink in Brackish Lake Akkeshi, Japan TATSUNORI NAKAGAWA1*, YUKI TSUCHIYA1, SHINGO UEDA1, MANABU FUKUI2, and REIJI TAKAHASHI1 1College of Bioresource Sciences, Nihon University, 1866 Kameino, Fujisawa, 252–0880, Japan; and 2Institute of Low Temperature Science, Hokkaido University, Kita-19, Nishi-8, Kita-ku, Sapporo, 060–0819, Japan (Received July 16, 2018—Accepted October 22, 2018—Published online December 1, 2018) Nitrous oxide (N2O) is a powerful greenhouse gas; however, limited information is currently available on the microbiomes involved in its sink and source in seagrass meadow sediments. Using laboratory incubations, a quantitative PCR (qPCR) analysis of N2O reductase (nosZ) and ammonia monooxygenase subunit A (amoA) genes, and a metagenome analysis based on the nosZ gene, we investigated the abundance of N2O-reducing microorganisms and ammonia-oxidizing prokaryotes as well as the community compositions of N2O-reducing microorganisms in in situ and cultivated sediments in the non-eelgrass and eelgrass zones of Lake Akkeshi, Japan. Laboratory incubations showed that N2O was reduced by eelgrass sediments and emitted by non-eelgrass sediments. qPCR analyses revealed that the abundance of nosZ gene clade II in both sediments before and after the incubation as higher in the eelgrass zone than in the non-eelgrass zone. In contrast, the abundance of ammonia-oxidizing archaeal amoA genes increased after incubations in the non-eelgrass zone only. Metagenome analyses of nosZ genes revealed that the lineages Dechloromonas-Magnetospirillum-Thiocapsa and Bacteroidetes (Flavobacteriia) within nosZ gene clade II were the main populations in the N2O-reducing microbiome in the in situ sediments of eelgrass zones. -
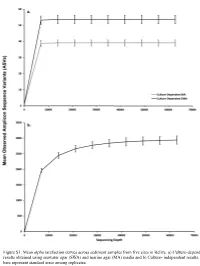
Figure S1. Mean Alpha Rarefaction Curves Across Sediment Samples from Five Sites in Belize
Figure S1. Mean alpha rarefaction curves across sediment samples from five sites in Belize. a) Culture-dependent results obtained using seawater agar (SWA) and marine agar (MA) media and b) Culture- independent results. Error bars represent standard error among replicates. a. 200 150 100 Faith’s PD Faith’s 50 0 Culturedependent MA Culturedependent SWA Cultureindependent Method b. 0.8 0.6 Pielou’s Evenness 0.4 Culturedependent MA Culturedependent SWA Cultureindependent Method Figure S2. Alpha diversity boxplots of marine sediment microbial communities from Carrie Bow Cay, Belize in culture-dependent and culture-independent samples determined using a) Faith’s Phylogenetic Diversity Index and b) Pielou’s Evenness. Culture-dependent methods include the use of marine agar medium (MA) and seawater agar medium (SWA. Data points are overlayed on the boxplot to show variation. a. 100% Proteobacteria Nitrospinae Bacteroidetes Fibrobacteres Planctomycetes Entotheonellaeota 90% Cyanobacteria Marinimicrobia (SAR406 clade) Firmicutes Deinococcus-Thermus Chloroflexi Margulisbacteria [A] Thaumarchaeota Unassigned 80% Acidobacteria Archaea UA Verrucomicrobia Acetothermia Actinobacteria Elusimicrobia 70% [A] Nanoarchaeaeota Tenericutes Kiritimatiellaeota TA06 Latescibacteria WS2 60% Spirochaetes Dependentiae Patescibacteria LCP-89 Gemmatimonadetes FCPU426 Omnitrophicaeota WPS-2 50% Fusobacteria CK-2C2-2 Bacteria UA Armatimonadetes [A] Euryarchaeota [A] Altiarchaeota Relative Percent 40% Calditrichaeota Poribacteria Lentisphaerae Cloacimonetes [A] Crenarchaeota -

DNA Variation and Symbiotic Associations in Phenotypically Diverse Sea Urchin Strongylocentrotus Intermedius
DNA variation and symbiotic associations in phenotypically diverse sea urchin Strongylocentrotus intermedius Evgeniy S. Balakirev*†‡, Vladimir A. Pavlyuchkov§, and Francisco J. Ayala*‡ *Department of Ecology and Evolutionary Biology, University of California, Irvine, CA 92697-2525; †Institute of Marine Biology, Vladivostok 690041, Russia; and §Pacific Research Fisheries Centre (TINRO-Centre), Vladivostok, 690600 Russia Contributed by Francisco J. Ayala, August 20, 2008 (sent for review May 9, 2008) Strongylocentrotus intermedius (A. Agassiz, 1863) is an economically spines of the U form are relatively short; the length, as a rule, does important sea urchin inhabiting the northwest Pacific region of Asia. not exceed one third of the radius of the testa. The spines of the G The northern Primorye (Sea of Japan) populations of S. intermedius form are longer, reaching and frequently exceeding two thirds of the consist of two sympatric morphological forms, ‘‘usual’’ (U) and ‘‘gray’’ testa radius. The testa is significantly thicker in the U form than in (G). The two forms are significantly different in morphology and the G form. The morphological differences between the U and G preferred bathymetric distribution, the G form prevailing in deeper- forms of S. intermedius are stable and easily recognizable (Fig. 1), water settlements. We have analyzed the genetic composition of the and they are systematically reported for the northern Primorye S. intermedius forms using the nucleotide sequences of the mitochon- coast region (V.A.P., unpublished data). drial gene encoding the cytochrome c oxidase subunit I and the Little is known about the population genetics of S. intermedius; nuclear gene encoding bindin to evaluate the possibility of cryptic the available data are limited to allozyme polymorphisms (4–6). -

08 Criobiology Rosario.P65
BIOCELL ISSN 0327 - 9545 2009, 33(3): A225-A241 PRINTED IN ARGENTINA Centro Binacional de Criobiología Clínica y Aplicada UNESCO Chair in Criobiology Facultad de Ciencias Bioquímicas y Farmacéuticas Universidad Nacional de Rosario Suipacha 570 (S2002LRK) Rosario Argentina Lectures and Abstracts from the 3er WORKSHOP IN CRYOBIOLOGY APPLIED TO MEDICAL SCIENCES May 5-7, 2009 Rosario, ARGENTINA The lectures and abstracts from 3er Workshop in Cryobiology of Medical Sciences have been revised and evaluated by a scientific committee A226 BIOCELL 33(3), 2009 Board of the 3er Workshop in Cryobiology Applied to Medical Sciences President Prof. Dr. Edgardo E. Guibert Vicepresident Prof. Dr. Joaquín V. Rodriguez Secretary Dra. Alejandra B. Quintana Sub-Secretary Dra. María Eugenia Mamprin Tresurer Dra. María Gabriela Mediavilla Collaborators Lic. María Dolores Pizarro Dra. Valeria Sigot Dra. Cecilia Mansilla Scientific Committee Dr. Blas Melissari (Uruguay) Dra. Alejandra Quintana (Argentina) Dra. María Eugenia Mamprin (Argentina) Dra. María Gabriela Mediavilla (Argentina) Dra. Valeria Sigot (Argentina) Governmental and Private sponsors ✓ Agencia Nacional de Promoción Científica y Tecnológica ✓ Secretaría de Ciencia, Tecnología e Innovación Productiva - SECYT ✓ Universidad Nacional de Rosario ✓ Gobierno de la Provincia de Santa Fe, Sub-Secretaría de Ciencia y Técnica ✓ UNESCO Chair in Cryobiology ✓ Cámara de Comercio Italiana de Rosario ✓ Consulado de Italia ✓ Centro Studi Fegato, Trieste, Italy ✓ AGA ✓ Claudio Lamelza S.R.L. ✓ Titania Viajes y Turismo -

Colwellia and Marinobacter Metapangenomes Reveal Species
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.28.317438; this version posted September 28, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Colwellia and Marinobacter metapangenomes reveal species-specific responses to oil 2 and dispersant exposure in deepsea microbial communities 3 4 Tito David Peña-Montenegro1,2,3, Sara Kleindienst4, Andrew E. Allen5,6, A. Murat 5 Eren7,8, John P. McCrow5, Juan David Sánchez-Calderón3, Jonathan Arnold2,9, Samantha 6 B. Joye1,* 7 8 Running title: Metapangenomes reveal species-specific responses 9 10 1 Department of Marine Sciences, University of Georgia, 325 Sanford Dr., Athens, 11 Georgia 30602-3636, USA 12 13 2 Institute of Bioinformatics, University of Georgia, 120 Green St., Athens, Georgia 14 30602-7229, USA 15 16 3 Grupo de Investigación en Gestión Ecológica y Agroindustrial (GEA), Programa de 17 Microbiología, Facultad de Ciencias Exactas y Naturales, Universidad Libre, Seccional 18 Barranquilla, Colombia 19 20 4 Microbial Ecology, Center for Applied Geosciences, University of Tübingen, 21 Schnarrenbergstrasse 94-96, 72076 Tübingen, Germany 22 23 5 Microbial and Environmental Genomics, J. Craig Venter Institute, La Jolla, CA 92037, 24 USA 25 26 6 Integrative Oceanography Division, Scripps Institution of Oceanography, UC San 27 Diego, La Jolla, CA 92037, USA 28 29 7 Department of Medicine, University of Chicago, Chicago, IL, USA 30 31 8 Josephine Bay Paul Center, Marine Biological Laboratory, Woods Hole, MA, USA 32 33 9Department of Genetics, University of Georgia, 120 Green St., Athens, Georgia 30602- 34 7223, USA 35 36 *Correspondence: Samantha B. -

Compile.Xlsx
Silva OTU GS1A % PS1B % Taxonomy_Silva_132 otu0001 0 0 2 0.05 Bacteria;Acidobacteria;Acidobacteria_un;Acidobacteria_un;Acidobacteria_un;Acidobacteria_un; otu0002 0 0 1 0.02 Bacteria;Acidobacteria;Acidobacteriia;Solibacterales;Solibacteraceae_(Subgroup_3);PAUC26f; otu0003 49 0.82 5 0.12 Bacteria;Acidobacteria;Aminicenantia;Aminicenantales;Aminicenantales_fa;Aminicenantales_ge; otu0004 1 0.02 7 0.17 Bacteria;Acidobacteria;AT-s3-28;AT-s3-28_or;AT-s3-28_fa;AT-s3-28_ge; otu0005 1 0.02 0 0 Bacteria;Acidobacteria;Blastocatellia_(Subgroup_4);Blastocatellales;Blastocatellaceae;Blastocatella; otu0006 0 0 2 0.05 Bacteria;Acidobacteria;Holophagae;Subgroup_7;Subgroup_7_fa;Subgroup_7_ge; otu0007 1 0.02 0 0 Bacteria;Acidobacteria;ODP1230B23.02;ODP1230B23.02_or;ODP1230B23.02_fa;ODP1230B23.02_ge; otu0008 1 0.02 15 0.36 Bacteria;Acidobacteria;Subgroup_17;Subgroup_17_or;Subgroup_17_fa;Subgroup_17_ge; otu0009 9 0.15 41 0.99 Bacteria;Acidobacteria;Subgroup_21;Subgroup_21_or;Subgroup_21_fa;Subgroup_21_ge; otu0010 5 0.08 50 1.21 Bacteria;Acidobacteria;Subgroup_22;Subgroup_22_or;Subgroup_22_fa;Subgroup_22_ge; otu0011 2 0.03 11 0.27 Bacteria;Acidobacteria;Subgroup_26;Subgroup_26_or;Subgroup_26_fa;Subgroup_26_ge; otu0012 0 0 1 0.02 Bacteria;Acidobacteria;Subgroup_5;Subgroup_5_or;Subgroup_5_fa;Subgroup_5_ge; otu0013 1 0.02 13 0.32 Bacteria;Acidobacteria;Subgroup_6;Subgroup_6_or;Subgroup_6_fa;Subgroup_6_ge; otu0014 0 0 1 0.02 Bacteria;Acidobacteria;Subgroup_6;Subgroup_6_un;Subgroup_6_un;Subgroup_6_un; otu0015 8 0.13 30 0.73 Bacteria;Acidobacteria;Subgroup_9;Subgroup_9_or;Subgroup_9_fa;Subgroup_9_ge; -

Halophilic Bacteroidetes As an Example on How Their Genomes Interact with the Environment
DOCTORAL THESIS 2020 PHYLOGENOMICS OF BACTEROIDETES; HALOPHILIC BACTEROIDETES AS AN EXAMPLE ON HOW THEIR GENOMES INTERACT WITH THE ENVIRONMENT Raúl Muñoz Jiménez DOCTORAL THESIS 2020 Doctoral Programme of Environmental and Biomedical Microbiology PHYLOGENOMICS OF BACTEROIDETES; HALOPHILIC BACTEROIDETES AS AN EXAMPLE ON HOW THEIR GENOMES INTERACT WITH THE ENVIRONMENT Raúl Muñoz Jiménez Thesis Supervisor: Ramon Rosselló Móra Thesis Supervisor: Rudolf Amann Thesis tutor: Elena I. García-Valdés Pukkits Doctor by the Universitat de les Illes Balears Publications resulted from this thesis Munoz, R., Rosselló-Móra, R., & Amann, R. (2016). Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov. Systematic and Applied Microbiology, 39(5), 281–296 Munoz, R., Rosselló-Móra, R., & Amann, R. (2016). Corrigendum to “Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov.” [Syst. Appl. Microbiol. 39 (5) (2016) 281–296]. Systematic and Applied Microbiology, 39, 491–492. Munoz, R., Amann, R., & Rosselló-Móra, R. (2019). Ancestry and adaptive radiation of Bacteroidetes as assessed by comparative genomics. Systematic and Applied Microbiology, 43(2), 126065. Dr. Ramon Rosselló Móra, of the Institut Mediterrani d’Estudis Avançats, Esporles and Dr. Rudolf Amann, of the Max-Planck-Institute für Marine Mikrobiologie, Bremen WE DECLARE: That the thesis titled Phylogenomics of Bacteroidetes; halophilic Bacteroidetes as an example on how their genomes interact with the environment, presented by Raúl Muñoz Jiménez to obtain a doctoral degree, has been completed under our supervision and meets the requirements to opt for an International Doctorate. For all intents and purposes, we hereby sign this document. -

Metagenomics Unveils the Attributes of the Alginolytic Guilds of Sediments from Four Distant Cold Coastal Environments
Metagenomics unveils the attributes of the alginolytic guilds of sediments from four distant cold coastal environments Marina N Matos1, Mariana Lozada1, Luciano E Anselmino1, Matías A Musumeci1, Bernard Henrissat2,3,4, Janet K Jansson5, Walter P Mac Cormack6,7, JoLynn Carroll8,9, Sara Sjöling10, Leif Lundgren11 and Hebe M Dionisi1* 1Laboratorio de Microbiología Ambiental, Centro para el Estudio de Sistemas Marinos (CESIMAR, CONICET), Puerto Madryn, U9120ACD, Chubut, Argentina 2Architecture et Fonction des Macromolécules Biologiques, CNRS, Aix-Marseille Université, 13288 Marseille, France 3INRA, USC 1408 AFMB, F-13288 Marseille, France 4Department of Biological Sciences, King Abdulaziz University, Jeddah, 21589, Saudi Arabia 5Earth and Biological Sciences Directorate, Pacific Northwest National Laboratory, Richland, WA 99352, USA 6Instituto Antártico Argentino, Ciudad Autónoma de Buenos Aires, C1064ABR, Argentina 7Instituto Nanobiotec, CONICET- Universidad de Buenos Aires, Ciudad Autónoma de Buenos Aires, C1113AAC, Argentina 8Akvaplan-niva, Fram – High North Research Centre for Climate and the Environment, NO- 9296 Tromsø, Norway 9CAGE - Centre for Arctic Gas Hydrate, Environment and Climate, UiT The Arctic University of Norway, N-9037 Tromsø, Norway 10School of Natural Sciences and Environmental Studies, Södertörn University, 141 89 Huddinge, Sweden 11Stockholm University, SE-106 91 Stockholm, Sweden This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process which may lead to differences between this version and the Version of Record. Please cite this article as an ‘Accepted Article’, doi: 10.1111/1462-2920.13433 This article is protected by copyright. All rights reserved. Page 2 of 41 Running title: Alginolytic guilds from cold sediments *Correspondence: Hebe M. -

Ichthyenterobacterium Magnum.Pdf
International Journal of Systematic and Evolutionary Microbiology (2015), 65, 1186–1192 DOI 10.1099/ijs.0.000078 Ichthyenterobacterium magnum gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from olive flounder (Paralichthys olivaceus) Qismat Shakeela, Ahmed Shehzad, Kaihao Tang, Yunhui Zhang and Xiao-Hua Zhang Correspondence College of Marine Life Sciences, Ocean University of China, Qingdao 266003, PR China Xiao-Hua Zhang [email protected] A novel marine bacterium isolated from the intestine of cultured flounder (Paralichthys olivaceus) was studied by using a polyphasic taxonomic approach. The isolate was Gram-stain-negative, pleomorphic, aerobic, yellow and oxidase- and catalase-negative. Phylogenetic analysis of 16S rRNA gene sequences indicated that isolate Th6T formed a distinct branch within the family Flavobacteriaceae and showed 96.6 % similarity to its closest relative, Bizionia hallyeonensis T- y7T. The DNA G+C content was 29 mol%. The major respiratory quinone was MK-6. The predominant fatty acids were iso-C15 : 1 G, iso-C15 : 0, iso-C15 : 0 3-OH, iso-C17 : 0 3-OH and summed feature 3 (C15 : 1v6c and/or C16 : 1v7c). On the basis of the phenotypic, chemotaxo- nomic and phylogenetic characteristics, the novel bacterium has been assigned to a novel species of a new genus for which the name Ichthyenterobacterium magnum gen. nov., sp. nov. is proposed. The type strain is Th6T (5JCM 18636T5KCTC 32140T). The family Flavobacteriaceae was proposed by Jooste sampled aseptically by dissecting the fish. Th6T was isolated (1985) and was included in the first edition of Bergey’s from the tissue homogenate by the plate spreading method Manual of Systematic Bacteriology (Reichenbach, 1989). -

The Intestinal Bacterial Community and Functional Potential of Litopenaeus Vannamei in the Coastal Areas of China
microorganisms Article The Intestinal Bacterial Community and Functional Potential of Litopenaeus vannamei in the Coastal Areas of China Yimeng Cheng 1, Chaorong Ge 1,*, Wei Li 1 and Huaiying Yao 1,2,3 1 Research Center for Environmental Ecology and Engineering, School of Environmental Ecology and Biological Engineering, Wuhan Institute of Technology, Wuhan 430073, China; [email protected] (Y.C.); [email protected] (W.L.); [email protected] (H.Y.) 2 Zhejiang Key Laboratory of Urban Environmental Processes and Pollution Control, Ningbo Urban Environment Observation and Research Station, Chinese Academy of Sciences, Ningbo 315800, China 3 Key Laboratory of Urban Environment and Health, Institute of Urban Environment, Chinese Academy of Sciences, Xiamen 361021, China * Correspondence: [email protected] Abstract: Intestinal bacteria are crucial for the healthy aquaculture of Litopenaeus vannamei, and the coastal areas of China are important areas for concentrated L. vannamei cultivation. In this study, we evaluated different compositions and structures, key roles, and functional potentials of the intestinal bacterial community of L. vannamei shrimp collected in 12 Chinese coastal cities and investigated the correlation between the intestinal bacteria and functional potentials. The dominant bacteria in the shrimp intestines included Proteobacteria, Bacteroidetes, Tenericutes, Firmicutes, and Actinobacteria, and the main potential functions were metabolism, genetic information processing, and environmental information processing. Although the composition and structure of the intestinal bacterial community, potential pathogenic bacteria, and spoilage organisms varied from region to region, the functional potentials were homeostatic and significantly (p < 0.05) correlated with Citation: Cheng, Y.; Ge, C.; Li, W.; intestinal bacteria (at the family level) to different degrees. -

Contents Topic 1. Introduction to Microbiology. the Subject and Tasks
Contents Topic 1. Introduction to microbiology. The subject and tasks of microbiology. A short historical essay………………………………………………………………5 Topic 2. Systematics and nomenclature of microorganisms……………………. 10 Topic 3. General characteristics of prokaryotic cells. Gram’s method ………...45 Topic 4. Principles of health protection and safety rules in the microbiological laboratory. Design, equipment, and working regimen of a microbiological laboratory………………………………………………………………………….162 Topic 5. Physiology of bacteria, fungi, viruses, mycoplasmas, rickettsia……...185 TOPIC 1. INTRODUCTION TO MICROBIOLOGY. THE SUBJECT AND TASKS OF MICROBIOLOGY. A SHORT HISTORICAL ESSAY. Contents 1. Subject, tasks and achievements of modern microbiology. 2. The role of microorganisms in human life. 3. Differentiation of microbiology in the industry. 4. Communication of microbiology with other sciences. 5. Periods in the development of microbiology. 6. The contribution of domestic scientists in the development of microbiology. 7. The value of microbiology in the system of training veterinarians. 8. Methods of studying microorganisms. Microbiology is a science, which study most shallow living creatures - microorganisms. Before inventing of microscope humanity was in dark about their existence. But during the centuries people could make use of processes vital activity of microbes for its needs. They could prepare a koumiss, alcohol, wine, vinegar, bread, and other products. During many centuries the nature of fermentations remained incomprehensible. Microbiology learns morphology, physiology, genetics and microorganisms systematization, their ecology and the other life forms. Specific Classes of Microorganisms Algae Protozoa Fungi (yeasts and molds) Bacteria Rickettsiae Viruses Prions The Microorganisms are extraordinarily widely spread in nature. They literally ubiquitous forward us from birth to our death. Daily, hourly we eat up thousands and thousands of microbes together with air, water, food. -

Centro De Investigación En Alimentación Y Desarrollo, A.C
Centro de Investigación en Alimentación y Desarrollo, A.C. CARACTERIZACIÓN DE LA COMUNIDAD BACTERIANA DEL CORAL POCILLOPORA CAPITATA DE LOS PARCHES CORALINOS DE MANZANILLO, COLIMA. Por: Lic. María de los Angeles Milagros Laurel Sandoval TESIS APROBADA POR LA: UNIDAD MAZATLÁN EN ACUICULTURA Y MANEJO AMBIENTAL Como requisito parcial para obtener el grado de MAESTRÍA EN CIENCIAS Mazatlán, Sinaloa. Enero, 2014 ii DECLARACIÓN INSTITUCIONAL La información generada en esta tesis es propiedad intelectual del Centro de Investigación en Alimentación y Desarrollo, A.C. Se permiten y agradecen las citas breves del material contenido en esta tesis sin permiso especial del autor, siempre y cuando se dé crédito correspondiente. Para la reproducción parcial o total de la tesis con fines académicos, se deberá contar con la autorización escrita del director del Centro de Investigación en Alimentación y Desarrollo, A.C. (CIAD). La publicación en comunicaciones científicas o de divulgación popular de los datos contenidos en esta tesis, deberá dar los créditos al CIAD, previa autorización escrita del manuscrito en cuestión del director de tesis. Dr. Pablo Wong González Director General iii AGRADECIEMIENTOS Al Consejo Nacional de Ciencia y Tecnología (CONACyT) por el apoyo económico brindado durante mi maestría en el CIAD, así como también por la beca mixta asignada para la realización de la estancia en en la Universidad del Estado de San Diego (SDSU). Al Centro de Investigación en Alimentación y Desarrollo (CIAD), por permitirme la oportunidad de llevar a cabo mis estudios de maestría en su programa de posgrado. A mi directora de tesis la Dra. Sonia Araceli Soto Rodríguez por su confianza y apoyo en la realización de esta tesis, agradeciéndole sus aportaciones y tiempo para alcanzar esta meta.