Bacterial Taxa Based on Greengenes Database GS1A PS1B ABY1 OD1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

A Putative Cystathionine Beta-Synthase Homolog of Mycolicibacterium Smegmatis Is Involved in De Novo Cysteine Biosynthesis
University of Arkansas, Fayetteville ScholarWorks@UARK Theses and Dissertations 5-2020 A Putative Cystathionine Beta-Synthase Homolog of Mycolicibacterium smegmatis is Involved in de novo Cysteine Biosynthesis Saroj Kumar Mahato University of Arkansas, Fayetteville Follow this and additional works at: https://scholarworks.uark.edu/etd Part of the Cell Biology Commons, Molecular Biology Commons, and the Pathogenic Microbiology Commons Citation Mahato, S. K. (2020). A Putative Cystathionine Beta-Synthase Homolog of Mycolicibacterium smegmatis is Involved in de novo Cysteine Biosynthesis. Theses and Dissertations Retrieved from https://scholarworks.uark.edu/etd/3639 This Thesis is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of ScholarWorks@UARK. For more information, please contact [email protected]. A Putative Cystathionine Beta-Synthase Homolog of Mycolicibacterium smegmatis is Involved in de novo Cysteine Biosynthesis A thesis submitted in partial fulfillment of the requirement for the degree of Master of Science in Cell and Molecular Biology by Saroj Kumar Mahato Purbanchal University Bachelor of Science in Biotechnology, 2016 May 2020 University of Arkansas This thesis is approved for recommendation to the Graduate Council. ___________________________________ Young Min Kwon, Ph.D. Thesis Director ___________________________________ ___________________________________ Suresh Thallapuranam, Ph.D. Inés Pinto, Ph.D. Committee Member Committee Member ABSTRACT Mycobacteria include serious pathogens of humans and animals. Mycolicibacterium smegmatis is a non-pathogenic model that is widely used to study core mycobacterial metabolism. This thesis explores mycobacterial pathways of cysteine biosynthesis by generating and study of genetic mutants of M. smegmatis. Published in vitro biochemical studies had revealed three independent routes to cysteine synthesis in mycobacteria involving separate homologs of cysteine synthase, namely CysK1, CysK2, and CysM. -

Microbial Community Structure Dynamics in Ohio River Sediments During Reductive Dechlorination of Pcbs
University of Kentucky UKnowledge University of Kentucky Doctoral Dissertations Graduate School 2008 MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS Andres Enrique Nunez University of Kentucky Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Nunez, Andres Enrique, "MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS" (2008). University of Kentucky Doctoral Dissertations. 679. https://uknowledge.uky.edu/gradschool_diss/679 This Dissertation is brought to you for free and open access by the Graduate School at UKnowledge. It has been accepted for inclusion in University of Kentucky Doctoral Dissertations by an authorized administrator of UKnowledge. For more information, please contact [email protected]. ABSTRACT OF DISSERTATION Andres Enrique Nunez The Graduate School University of Kentucky 2008 MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS ABSTRACT OF DISSERTATION A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the College of Agriculture at the University of Kentucky By Andres Enrique Nunez Director: Dr. Elisa M. D’Angelo Lexington, KY 2008 Copyright © Andres Enrique Nunez 2008 ABSTRACT OF DISSERTATION MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS The entire stretch of the Ohio River is under fish consumption advisories due to contamination with polychlorinated biphenyls (PCBs). In this study, natural attenuation and biostimulation of PCBs and microbial communities responsible for PCB transformations were investigated in Ohio River sediments. Natural attenuation of PCBs was negligible in sediments, which was likely attributed to low temperature conditions during most of the year, as well as low amounts of available nitrogen, phosphorus, and organic carbon. -

Description of New and Amended Clades of the Genus Photobacterium
microorganisms Article Description of New and Amended Clades of the Genus Photobacterium Alejandro M. Labella 1, M. Dolores Castro 1 ID , Manuel Manchado 2 ID and Juan J. Borrego 1,* ID 1 Department of Microbiology, Faculty of Sciences, Universidad de Malaga, 29071 Malaga, Spain; [email protected] (A.M.L.); [email protected] (M.D.C.) 2 Puerto de Santa María, Junta de Andalucía, IFAPA Centro El Toruño, 11500 Cadiz, Spain; [email protected] * Correspondence: [email protected]; Tel.: +34-952131893 Received: 15 January 2018; Accepted: 7 March 2018; Published: 12 March 2018 Abstract: Phylogenetic relationships between species in the genus Photobacterium have been poorly studied despite pathogenic and ecological relevance of some of its members. This is the first phylogenetic study that includes new species of Photobacterium (validated or not) that have not been included in any of the previously described clades, using 16S rRNA sequences and multilocus sequence analysis (MLSA) in concatenated sequences of gyrB, gapA, topA, ftsZ and mreB housekeeping genes. Sequence analysis has been implemented using Maximum-parsimony (MP), Neighbour-joining (NJ) and Maximum likelihood (ML) treeing methods and the predicted evolutionary relationship between the Photobacterium clades was established on the basis of bootstrap values of >75% for 16S rRNA sequences and MLSA. We have grouped 22 species of the genus Photobacterium into the following 5 clades: Phosphoreum (comprises P. aquimaris,“P. carnosum,” P. iliopiscarium, P. kishitanii, P. phosphoreum,“P. piscicola” and “P. toruni”); clade Profundum (composed of P. aestuarii, P. alginatilyticum, P. frigidiphilum, P. indicum, P. jeanii, P. lipolyticum,“P. marinum,” and P. profundum); clade Damselae (two subspecies of P. -

Ameliorating Process Parameters for Zeaxanthin Yield in Arthrobacter Gandavensis MTCC 25325
Ram et al. AMB Expr (2020) 10:69 https://doi.org/10.1186/s13568-020-01008-4 ORIGINAL ARTICLE Open Access Ameliorating process parameters for zeaxanthin yield in Arthrobacter gandavensis MTCC 25325 Shristi Ram1,2, Sushma Rani Tirkey1,2, Madhava Anil Kumar1,3 and Sandhya Mishra1,2* Abstract The present study aims to escalate the production of prophylactic agent zeaxanthin using a screened potential bacterial isolate. For this purpose, a freshwater bacterium capable of producing zeaxanthin was isolated from Bor Talav, Bhavnagar. The 16S rRNA sequence confrmed the isolate as Arthrobacter gandavensis. The bacterium was also submitted to Microbial Type Culture Collection, CSIR-Institute of Microbial Technology, Chandigarh, India, with the accession number MTCC 25325. The chemo-metric tools were employed to optimise the infuencing factors such as pH, temperature, inoculum size, agitation speed, carbon source and harvest time on zeaxanthin yield. Thereafter, six parameters were narrowed down to three factors and were optimised using the central composite design (CCD) matrix. Maximum zeaxanthin (1.51 mg/g) was derived when A. gandavensis MTCC 25325 was grown under pH 6.0, 1.5% (w/v) glucose and 10% (v/v) inoculum size. A high regression coefcient (R2 0.92) of the developed model indicated the accurateness of the tested parameters. To the best of our knowledge,= this is the frst report on tailoring the process parameters using chemo-metric optimisation for escalating the zeaxanthin production by A. gandavensis MTCC 25325. Keywords: Arthrobacter gandavensis MTCC 25325, Central composite design, Nutraceutical, Zeaxanthin Key points Introduction Te pursuit of alternative sources as functional food (that • Chemo-metric approach was used for optimising serves the dual purpose of nutrition and diet) has led to zeaxanthin production. -

Pigments Produced by the Bacteria Belonging to the Genus Arthrobacter Nuthathai Sutthiwong, Yanis Caro, Mireille Fouillaud, Philippe Laurent, A
Pigments produced by the bacteria belonging to the genus Arthrobacter Nuthathai Sutthiwong, Yanis Caro, Mireille Fouillaud, Philippe Laurent, A. Valla, Laurent Dufossé To cite this version: Nuthathai Sutthiwong, Yanis Caro, Mireille Fouillaud, Philippe Laurent, A. Valla, et al.. Pigments produced by the bacteria belonging to the genus Arthrobacter. 7th International Congress of Pigments in Food – New technologies towards health, through colors, Jun 2013, Novara, Italy. 2016. hal- 01397507 HAL Id: hal-01397507 https://hal.archives-ouvertes.fr/hal-01397507 Submitted on 16 Nov 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Public Domain Pigments produced by the bacteria belonging to the genus Arthrobacter Sutthiwong N.1, 2, Caro Y.2, Fouillaud M.2, Laurent P.3, Valla A.4, Dufossé L.2,5 1 Agricultural Technology Department, Thailand Institute of Scientific and Technological Research, Pathum Thani, Thailand 2 Laboratoire de Chimie des Substances Naturelles et des Sciences des Aliments, Université de La Réunion, ESIROI Agroalimentaire,Sainte-Clotilde, -

Alpine Soil Bacterial Community and Environmental Filters Bahar Shahnavaz
Alpine soil bacterial community and environmental filters Bahar Shahnavaz To cite this version: Bahar Shahnavaz. Alpine soil bacterial community and environmental filters. Other [q-bio.OT]. Université Joseph-Fourier - Grenoble I, 2009. English. tel-00515414 HAL Id: tel-00515414 https://tel.archives-ouvertes.fr/tel-00515414 Submitted on 6 Sep 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE Pour l’obtention du titre de l'Université Joseph-Fourier - Grenoble 1 École Doctorale : Chimie et Sciences du Vivant Spécialité : Biodiversité, Écologie, Environnement Communautés bactériennes de sols alpins et filtres environnementaux Par Bahar SHAHNAVAZ Soutenue devant jury le 25 Septembre 2009 Composition du jury Dr. Thierry HEULIN Rapporteur Dr. Christian JEANTHON Rapporteur Dr. Sylvie NAZARET Examinateur Dr. Jean MARTIN Examinateur Dr. Yves JOUANNEAU Président du jury Dr. Roberto GEREMIA Directeur de thèse Thèse préparée au sien du Laboratoire d’Ecologie Alpine (LECA, UMR UJF- CNRS 5553) THÈSE Pour l’obtention du titre de Docteur de l’Université de Grenoble École Doctorale : Chimie et Sciences du Vivant Spécialité : Biodiversité, Écologie, Environnement Communautés bactériennes de sols alpins et filtres environnementaux Bahar SHAHNAVAZ Directeur : Roberto GEREMIA Soutenue devant jury le 25 Septembre 2009 Composition du jury Dr. -
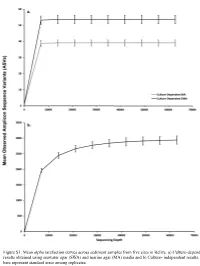
Figure S1. Mean Alpha Rarefaction Curves Across Sediment Samples from Five Sites in Belize
Figure S1. Mean alpha rarefaction curves across sediment samples from five sites in Belize. a) Culture-dependent results obtained using seawater agar (SWA) and marine agar (MA) media and b) Culture- independent results. Error bars represent standard error among replicates. a. 200 150 100 Faith’s PD Faith’s 50 0 Culturedependent MA Culturedependent SWA Cultureindependent Method b. 0.8 0.6 Pielou’s Evenness 0.4 Culturedependent MA Culturedependent SWA Cultureindependent Method Figure S2. Alpha diversity boxplots of marine sediment microbial communities from Carrie Bow Cay, Belize in culture-dependent and culture-independent samples determined using a) Faith’s Phylogenetic Diversity Index and b) Pielou’s Evenness. Culture-dependent methods include the use of marine agar medium (MA) and seawater agar medium (SWA. Data points are overlayed on the boxplot to show variation. a. 100% Proteobacteria Nitrospinae Bacteroidetes Fibrobacteres Planctomycetes Entotheonellaeota 90% Cyanobacteria Marinimicrobia (SAR406 clade) Firmicutes Deinococcus-Thermus Chloroflexi Margulisbacteria [A] Thaumarchaeota Unassigned 80% Acidobacteria Archaea UA Verrucomicrobia Acetothermia Actinobacteria Elusimicrobia 70% [A] Nanoarchaeaeota Tenericutes Kiritimatiellaeota TA06 Latescibacteria WS2 60% Spirochaetes Dependentiae Patescibacteria LCP-89 Gemmatimonadetes FCPU426 Omnitrophicaeota WPS-2 50% Fusobacteria CK-2C2-2 Bacteria UA Armatimonadetes [A] Euryarchaeota [A] Altiarchaeota Relative Percent 40% Calditrichaeota Poribacteria Lentisphaerae Cloacimonetes [A] Crenarchaeota -

Supplementary Information for Microbial Electrochemical Systems Outperform Fixed-Bed Biofilters for Cleaning-Up Urban Wastewater
Electronic Supplementary Material (ESI) for Environmental Science: Water Research & Technology. This journal is © The Royal Society of Chemistry 2016 Supplementary information for Microbial Electrochemical Systems outperform fixed-bed biofilters for cleaning-up urban wastewater AUTHORS: Arantxa Aguirre-Sierraa, Tristano Bacchetti De Gregorisb, Antonio Berná, Juan José Salasc, Carlos Aragónc, Abraham Esteve-Núñezab* Fig.1S Total nitrogen (A), ammonia (B) and nitrate (C) influent and effluent average values of the coke and the gravel biofilters. Error bars represent 95% confidence interval. Fig. 2S Influent and effluent COD (A) and BOD5 (B) average values of the hybrid biofilter and the hybrid polarized biofilter. Error bars represent 95% confidence interval. Fig. 3S Redox potential measured in the coke and the gravel biofilters Fig. 4S Rarefaction curves calculated for each sample based on the OTU computations. Fig. 5S Correspondence analysis biplot of classes’ distribution from pyrosequencing analysis. Fig. 6S. Relative abundance of classes of the category ‘other’ at class level. Table 1S Influent pre-treated wastewater and effluents characteristics. Averages ± SD HRT (d) 4.0 3.4 1.7 0.8 0.5 Influent COD (mg L-1) 246 ± 114 330 ± 107 457 ± 92 318 ± 143 393 ± 101 -1 BOD5 (mg L ) 136 ± 86 235 ± 36 268 ± 81 176 ± 127 213 ± 112 TN (mg L-1) 45.0 ± 17.4 60.6 ± 7.5 57.7 ± 3.9 43.7 ± 16.5 54.8 ± 10.1 -1 NH4-N (mg L ) 32.7 ± 18.7 51.6 ± 6.5 49.0 ± 2.3 36.6 ± 15.9 47.0 ± 8.8 -1 NO3-N (mg L ) 2.3 ± 3.6 1.0 ± 1.6 0.8 ± 0.6 1.5 ± 2.0 0.9 ± 0.6 TP (mg -

Distinct Bacterial Communities Associated with the Coral Model Aiptasia in Aposymbiotic and Symbiotic States with Symbiodinium
ORIGINAL RESEARCH published: 18 November 2016 doi: 10.3389/fmars.2016.00234 Distinct Bacterial Communities Associated with the Coral Model Aiptasia in Aposymbiotic and Symbiotic States with Symbiodinium Till Röthig †, Rúben M. Costa †, Fabia Simona, Sebastian Baumgarten, Ana F. Torres, Anand Radhakrishnan, Manuel Aranda and Christian R. Voolstra * Division of Biological and Environmental Science and Engineering (BESE), Red Sea Research Center, King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia Coral reefs are in decline. The basic functional unit of coral reefs is the coral metaorganism or holobiont consisting of the cnidarian host animal, symbiotic algae of the genus Symbiodinium, and a specific consortium of bacteria (among others), but research is slow due to the difficulty of working with corals. Aiptasia has proven to be a tractable Edited by: model system to elucidate the intricacies of cnidarian-dinoflagellate symbioses, but Thomas Carl Bosch, characterization of the associated bacterial microbiome is required to provide a complete University of Kiel, Germany and integrated understanding of holobiont function. In this work, we characterize and Reviewed by: analyze the microbiome of aposymbiotic and symbiotic Aiptasia and show that bacterial Simon K. Davy, Victoria University of Wellington, associates are distinct in both conditions. We further show that key microbial associates New Zealand can be cultured without their cnidarian host. Our results suggest that bacteria play an Mathieu Pernice, University of Technology, Australia important role in the symbiosis of Aiptasia with Symbiodinium, a finding that underlines *Correspondence: the power of the Aiptasia model system where cnidarian hosts can be analyzed in Christian R. Voolstra aposymbiotic and symbiotic states. -

Taxonomy JN869023
Species that differentiate periods of high vs. low species richness in unattached communities Species Taxonomy JN869023 Bacteria; Actinobacteria; Actinobacteria; Actinomycetales; ACK-M1 JN674641 Bacteria; Bacteroidetes; [Saprospirae]; [Saprospirales]; Chitinophagaceae; Sediminibacterium JN869030 Bacteria; Actinobacteria; Actinobacteria; Actinomycetales; ACK-M1 U51104 Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Limnohabitans JN868812 Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae JN391888 Bacteria; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Planctomyces HM856408 Bacteria; Planctomycetes; Phycisphaerae; Phycisphaerales GQ347385 Bacteria; Verrucomicrobia; [Methylacidiphilae]; Methylacidiphilales; LD19 GU305856 Bacteria; Proteobacteria; Alphaproteobacteria; Rickettsiales; Pelagibacteraceae GQ340302 Bacteria; Actinobacteria; Actinobacteria; Actinomycetales JN869125 Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae New.ReferenceOTU470 Bacteria; Cyanobacteria; ML635J-21 JN679119 Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae HM141858 Bacteria; Acidobacteria; Holophagae; Holophagales; Holophagaceae; Geothrix FQ659340 Bacteria; Verrucomicrobia; [Pedosphaerae]; [Pedosphaerales]; auto67_4W AY133074 Bacteria; Elusimicrobia; Elusimicrobia; Elusimicrobiales FJ800541 Bacteria; Verrucomicrobia; [Pedosphaerae]; [Pedosphaerales]; R4-41B JQ346769 Bacteria; Acidobacteria; [Chloracidobacteria]; RB41; Ellin6075 -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341