The Microbiome of the Egyptian Red Sea Proper and Gulf of Aqaba
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metaproteogenomic Insights Beyond Bacterial Response to Naphthalene
ORIGINAL ARTICLE ISME Journal – Original article Metaproteogenomic insights beyond bacterial response to 5 naphthalene exposure and bio-stimulation María-Eugenia Guazzaroni, Florian-Alexander Herbst, Iván Lores, Javier Tamames, Ana Isabel Peláez, Nieves López-Cortés, María Alcaide, Mercedes V. del Pozo, José María Vieites, Martin von Bergen, José Luis R. Gallego, Rafael Bargiela, Arantxa López-López, Dietmar H. Pieper, Ramón Rosselló-Móra, Jesús Sánchez, Jana Seifert and Manuel Ferrer 10 Supporting Online Material includes Text (Supporting Materials and Methods) Tables S1 to S9 Figures S1 to S7 1 SUPPORTING TEXT Supporting Materials and Methods Soil characterisation Soil pH was measured in a suspension of soil and water (1:2.5) with a glass electrode, and 5 electrical conductivity was measured in the same extract (diluted 1:5). Primary soil characteristics were determined using standard techniques, such as dichromate oxidation (organic matter content), the Kjeldahl method (nitrogen content), the Olsen method (phosphorus content) and a Bernard calcimeter (carbonate content). The Bouyoucos Densimetry method was used to establish textural data. Exchangeable cations (Ca, Mg, K and 10 Na) extracted with 1 M NH 4Cl and exchangeable aluminium extracted with 1 M KCl were determined using atomic absorption/emission spectrophotometry with an AA200 PerkinElmer analyser. The effective cation exchange capacity (ECEC) was calculated as the sum of the values of the last two measurements (sum of the exchangeable cations and the exchangeable Al). Analyses were performed immediately after sampling. 15 Hydrocarbon analysis Extraction (5 g of sample N and Nbs) was performed with dichloromethane:acetone (1:1) using a Soxtherm extraction apparatus (Gerhardt GmbH & Co. -

Extremozymes of the Hot and Salty Halothermothrix Orenii
Extremozymes of the Hot and Salty Halothermothrix orenii Author Kori, Lokesh D Published 2012 Thesis Type Thesis (PhD Doctorate) School School of Biomolecular and Physical Sciences DOI https://doi.org/10.25904/1912/2191 Copyright Statement The author owns the copyright in this thesis, unless stated otherwise. Downloaded from http://hdl.handle.net/10072/366220 Griffith Research Online https://research-repository.griffith.edu.au Extremozymes of the hot and salty Halothermothrix orenii LOKESH D. KORI (M.Sc. Biotechnology) School of Biomolecular and Physical Sciences Science, Environment, Engineering and Technology Griffith University, Australia Submitted in fulfillment of the requirements of the degree of Doctor of Philosophy December 2011 STATEMENT OF ORIGINALITY STATEMENT OF ORIGINALITY This work has not previously been submitted for a degree or diploma in any university. To the best of my knowledge and belief, the thesis contains no material previously published or written by another person except where due reference is made in the thesis itself. LOKESH DULICHAND KORI II ACKNOWLEDGEMENTS ACKNOWLEDGEMENTS I owe my deepest gratitude to my supervisor Prof. Bharat Patel, for offering me an opportunity for being his postgraduate. His boundless knowledge motivates me for keep going and enjoy the essence of science. Without his guidance, great patience and advice, I could not finish my PhD program successfully. I take this opportunity to give my heartiest thanks to Assoc. Prof. Andreas Hofmann, (Structural Chemistry, Eskitis Institute for Cell & Molecular Therapies, Griffith University) for his support and encouragement for crystallographic work. I am grateful to him for teaching me about the protein structures, in silico analysis and their hidden chemistry. -

METABOLIC EVOLUTION in GALDIERIA SULPHURARIA By
METABOLIC EVOLUTION IN GALDIERIA SULPHURARIA By CHAD M. TERNES Bachelor of Science in Botany Oklahoma State University Stillwater, Oklahoma 2009 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY May, 2015 METABOLIC EVOLUTION IN GALDIERIA SUPHURARIA Dissertation Approved: Dr. Gerald Schoenknecht Dissertation Adviser Dr. David Meinke Dr. Andrew Doust Dr. Patricia Canaan ii Name: CHAD M. TERNES Date of Degree: MAY, 2015 Title of Study: METABOLIC EVOLUTION IN GALDIERIA SULPHURARIA Major Field: PLANT SCIENCE Abstract: The thermoacidophilic, unicellular, red alga Galdieria sulphuraria possesses characteristics, including salt and heavy metal tolerance, unsurpassed by any other alga. Like most plastid bearing eukaryotes, G. sulphuraria can grow photoautotrophically. Additionally, it can also grow solely as a heterotroph, which results in the cessation of photosynthetic pigment biosynthesis. The ability to grow heterotrophically is likely correlated with G. sulphuraria ’s broad capacity for carbon metabolism, which rivals that of fungi. Annotation of the metabolic pathways encoded by the genome of G. sulphuraria revealed several pathways that are uncharacteristic for plants and algae, even red algae. Phylogenetic analyses of the enzymes underlying the metabolic pathways suggest multiple instances of horizontal gene transfer, in addition to endosymbiotic gene transfer and conservation through ancestry. Although some metabolic pathways as a whole appear to be retained through ancestry, genes encoding individual enzymes within a pathway were substituted by genes that were acquired horizontally from other domains of life. Thus, metabolic pathways in G. sulphuraria appear to be composed of a ‘metabolic patchwork’, underscored by a mosaic of genes resulting from multiple evolutionary processes. -

Genomic Insight Into the Host–Endosymbiont Relationship of Endozoicomonas Montiporae CL-33T with Its Coral Host
ORIGINAL RESEARCH published: 08 March 2016 doi: 10.3389/fmicb.2016.00251 Genomic Insight into the Host–Endosymbiont Relationship of Endozoicomonas montiporae CL-33T with its Coral Host Jiun-Yan Ding 1, Jia-Ho Shiu 1, Wen-Ming Chen 2, Yin-Ru Chiang 1 and Sen-Lin Tang 1* 1 Biodiversity Research Center, Academia Sinica, Taipei, Taiwan, 2 Department of Seafood Science, Laboratory of Microbiology, National Kaohsiung Marine University, Kaohsiung, Taiwan The bacterial genus Endozoicomonas was commonly detected in healthy corals in many coral-associated bacteria studies in the past decade. Although, it is likely to be a core member of coral microbiota, little is known about its ecological roles. To decipher potential interactions between bacteria and their coral hosts, we sequenced and investigated the first culturable endozoicomonal bacterium from coral, the E. montiporae CL-33T. Its genome had potential sign of ongoing genome erosion and gene exchange with its Edited by: Rekha Seshadri, host. Testosterone degradation and type III secretion system are commonly present in Department of Energy Joint Genome Endozoicomonas and may have roles to recognize and deliver effectors to their hosts. Institute, USA Moreover, genes of eukaryotic ephrin ligand B2 are present in its genome; presumably, Reviewed by: this bacterium could move into coral cells via endocytosis after binding to coral’s Eph Kathleen M. Morrow, University of New Hampshire, USA receptors. In addition, 7,8-dihydro-8-oxoguanine triphosphatase and isocitrate lyase Jean-Baptiste Raina, are possible type III secretion effectors that might help coral to prevent mitochondrial University of Technology Sydney, Australia dysfunction and promote gluconeogenesis, especially under stress conditions. -

Updating the Taxonomic Toolbox: Classification of Alteromonas Spp
1 Updating the taxonomic toolbox: classification of Alteromonas spp. 2 using Multilocus Phylogenetic Analysis and MALDI-TOF Mass 3 Spectrometry a a a 4 Hooi Jun Ng , Hayden K. Webb , Russell J. Crawford , François a b b c 5 Malherbe , Henry Butt , Rachel Knight , Valery V. Mikhailov and a, 6 Elena P. Ivanova * 7 aFaculty of Life and Social Sciences, Swinburne University of Technology, 8 PO Box 218, Hawthorn, Vic 3122, Australia 9 bBioscreen, Bio21 Institute, The University of Melbourne, Vic 3010, Australia 10 cG.B. Elyakov Pacific Institute of Bioorganic Chemistry, Far Eastern Branch, Russian 11 Academy of Sciences, Vladivostok 690022, Russian Federation 12 13 *Corresponding author: Tel: +61-3-9214-5137. Fax: +61-3-9214-5050. 14 E-mail: [email protected] 15 16 Abstract 17 Bacteria of the genus Alteromonas are Gram-negative, strictly aerobic, motile, 18 heterotrophic marine bacteria, known for their versatile metabolic activities. 19 Identification and classification of novel species belonging to the genus Alteromonas 20 generally involves DNA-DNA hybridization (DDH) as distinct species often fail to be 1 21 resolved at the 97% threshold value of the 16S rRNA gene sequence similarity. In this 22 study, the applicability of Multilocus Phylogenetic Analysis (MLPA) and Matrix- 23 Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF 24 MS) for the differentiation of Alteromonas species has been evaluated. Phylogenetic 25 analysis incorporating five house-keeping genes (dnaK, sucC, rpoB, gyrB, and rpoD) 26 revealed a threshold value of 98.9% that could be considered as the species cut-off 27 value for the delineation of Alteromonas spp. -

Granulicella Tundricola Type Strain MP5ACTX9T, an Acidobacteria from Tundra Soil
Standards in Genomic Sciences (2014) 9:449-461 DOI:10.4056/sig s.4648353 Complete genome sequence of Granulicella tundricola type strain MP5ACTX9T, an Acidobacteria from tundra soil Suman R. Rawat1, Minna K. Männistö 2, Valentin Starovoytov3, Lynne Goodwin4, Matt Nolan5 Loren Hauser6, Miriam Land 6, Karen Walston Davenport4, Tanja Woyke5 and Max M. Häggblom1* 1 Department of Biochemistry and Microbiology, Rutgers, The State University of New Jersey, New Brunswick, New Jersey USA 2 Finnish Forest Research Institute, Rovaniemi, Finland 3 Department of Cell Biology and Neuroscience, Rutgers, The State University of New Jersey, Piscataway, New Jersey, USA. 4 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA 5 DOE Joint Genome Institute, Walnut Creek, California, USA 6 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA *Correspondence: Max M Häggblom ([email protected]) Keywords: cold adapted, acidophile, tundra soil, Acidobacteria Granulicella tundricola strain MP5ACTX9T is a novel species of the genus Granulicella in subdivision 1 Acidobacteria. G. tundricola is a predominant member of soil bacterial communities, active at low temperatures and nutrient limiting conditions in Arctic alpine tundra. The organism is a cold-adapted acidophile and a versatile heterotroph that hydro- lyzes a suite of sugars and complex polysaccharides. Genome analysis revealed metabolic versatility with genes involved in metabolism and transport of carbohydrates, including g ene modules encoding for the carbohydrate-active enzyme (CAZy) families for the break- down, utilization and biosy nthesis of diverse structural and storag e polysaccharides suc h as plant based carbon polymers. The g enome of G. tundricola strain MP5ACTX9T consists of 4,309,151 bp of a circular chromosome and five meg a plasmids with a total genome con- tent of 5,503,984 bp. -

Microbiome Exploration of Deep-Sea Carnivorous Cladorhizidae Sponges
Microbiome exploration of deep-sea carnivorous Cladorhizidae sponges by Joost Theo Petra Verhoeven A Thesis submitted to the School of Graduate Studies in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Biology Memorial University of Newfoundland March 2019 St. John’s, Newfoundland and Labrador ABSTRACT Members of the sponge family Cladorhizidae are unique in having replaced the typical filter-feeding strategy of sponges by a predatory lifestyle, capturing and digesting small prey. These carnivorous sponges are found in many different environments, but are particularly abundant in deep waters, where they constitute a substantial component of the benthos. Sponges are known to host a wide range of microbial associates (microbiome) important for host health, but the extent of the microbiome in carnivorous sponges has never been extensively investigated and their importance is poorly understood. In this thesis, the microbiome of two deep-sea carnivorous sponge species (Chondrocladia grandis and Cladorhiza oxeata) is investigated for the first time, leveraging recent advances in high-throughput sequencing and through custom developed bioinformatic and molecular methods. Microbiome analyses showed that the carnivorous sponges co-occur with microorganisms and large differences in the composition and type of associations were observed between sponge species. Tissues of C. grandis hosted diverse bacterial communities, similar in composition between individuals, in stark contrast to C. oxeata where low microbial diversity was found with a high host-to-host variability. In C. grandis the microbiome was not homogeneous throughout the host tissue, and significant shifts occured within community members across anatomical regions, with the enrichment of specific bacterial taxa in particular anatomical niches, indicating a potential symbiotic role of such taxa within processes like prey digestion and chemolithoautotrophy. -
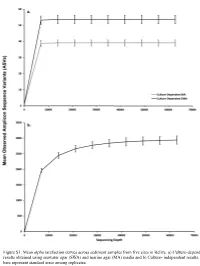
Figure S1. Mean Alpha Rarefaction Curves Across Sediment Samples from Five Sites in Belize
Figure S1. Mean alpha rarefaction curves across sediment samples from five sites in Belize. a) Culture-dependent results obtained using seawater agar (SWA) and marine agar (MA) media and b) Culture- independent results. Error bars represent standard error among replicates. a. 200 150 100 Faith’s PD Faith’s 50 0 Culturedependent MA Culturedependent SWA Cultureindependent Method b. 0.8 0.6 Pielou’s Evenness 0.4 Culturedependent MA Culturedependent SWA Cultureindependent Method Figure S2. Alpha diversity boxplots of marine sediment microbial communities from Carrie Bow Cay, Belize in culture-dependent and culture-independent samples determined using a) Faith’s Phylogenetic Diversity Index and b) Pielou’s Evenness. Culture-dependent methods include the use of marine agar medium (MA) and seawater agar medium (SWA. Data points are overlayed on the boxplot to show variation. a. 100% Proteobacteria Nitrospinae Bacteroidetes Fibrobacteres Planctomycetes Entotheonellaeota 90% Cyanobacteria Marinimicrobia (SAR406 clade) Firmicutes Deinococcus-Thermus Chloroflexi Margulisbacteria [A] Thaumarchaeota Unassigned 80% Acidobacteria Archaea UA Verrucomicrobia Acetothermia Actinobacteria Elusimicrobia 70% [A] Nanoarchaeaeota Tenericutes Kiritimatiellaeota TA06 Latescibacteria WS2 60% Spirochaetes Dependentiae Patescibacteria LCP-89 Gemmatimonadetes FCPU426 Omnitrophicaeota WPS-2 50% Fusobacteria CK-2C2-2 Bacteria UA Armatimonadetes [A] Euryarchaeota [A] Altiarchaeota Relative Percent 40% Calditrichaeota Poribacteria Lentisphaerae Cloacimonetes [A] Crenarchaeota -

Redalyc.Shallow-Water Hydrothermal Vents in the Azores (Portugal)
Revista de Gestão Costeira Integrada - Journal of Integrated Coastal Zone Management E-ISSN: 1646-8872 [email protected] Associação Portuguesa dos Recursos Hídricos Portugal Couto, Ruben P.; Rodriguesa, Armindo S.; Neto, Ana I. Shallow-water hydrothermal vents in the Azores (Portugal) Revista de Gestão Costeira Integrada - Journal of Integrated Coastal Zone Management, vol. 15, núm. 4, 2015, pp. 495-505 Associação Portuguesa dos Recursos Hídricos Lisboa, Portugal Available in: http://www.redalyc.org/articulo.oa?id=388343047005 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Revista de Gestão Costeira Integrada / Journal of Integrated Coastal Zone Management, 15(4):495-505 (2015) http://www.aprh.pt/rgci/pdf/rgci-584_Couto.pdf | DOI: 10.5894/rgci584 Shallow-water hydrothermal vents in the Azores (Portugal)* @, Ruben P. Couto@, a, b; Armindo S. Rodriguesa, c; Ana I. Netoa, d ABSTRACT The impact of global warming has been a major issue in recent years and will continue increasing in the future. Knowledge about the effects of ocean acidification on marine organisms and communities is crucial to efficient management. Island envi- ronments are particularly sensitive to externally induced changes and highly dependent on their coastal areas. This study summarises the published information on shallow-water hydrothermal vents of the Azores. These environments were reported to exhibit high metal concentration and acidified seawater due to the diffusion of acidic volcanic gases (mainly CO2) and a considerable temperature range. -

Impacts of Biogenic Polyunsaturated Aldehydes on Metabolism and Community
https://doi.org/10.5194/bg-2020-243 Preprint. Discussion started: 24 July 2020 c Author(s) 2020. CC BY 4.0 License. 1 Impacts of biogenic polyunsaturated aldehydes on metabolism and community 2 composition of particle-attached bacteria in coastal hypoxia 3 Zhengchao Wu1,2, Qian P. Li1,2,3,*, Zaiming Ge1,3, Bangqin Huang4, Chunming Dong5 4 1State Key Laboratory of Tropical Oceanography, South China Sea Institute of Oceanology, Chinese 5 Academy of Sciences, Guangzhou, China 6 2Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou, China 7 3College of Marine Science, University of the Chinese Academy of Sciences, Beijing, China 8 4Fujian Provincial Key Laboratory of Coastal Ecology and Environmental Studies, State Key Laboratory of 9 Marine Environmental Science, Xiamen University, Xiamen, China 10 5Key Laboratory of Marine Genetic Resources, Third Institute of Oceanography, MNR, Xiamen, China 11 *Correspondence to: Qian Li ([email protected]) 12 13 Abstract. Eutrophication-driven coastal hypoxia is of great interest recently, though its mechanisms are not 14 fully understood. Here, we showed elevated concentrations of particulate and dissolved polyunsaturated 15 aldehydes (PUAs) associated with the hypoxic waters meanly dominated by particle-attached bacteria (PAB) 16 in the bottom water of a salt-wedge estuary. Particle-adsorbed PUAs of ~10 micromoles per liter particle in 17 the hypoxic waters were directly quantified for the first time using large-volume-filtration followed with 18 on-site derivation and extraction of the adsorbed PUAs. PUAs-amended incubation experiments for PAB 19 retrieved from the low-oxygen waters were also performed to explore the impacts of PUAs on the growth 20 and metabolism of PAB and associated oxygen utilization. -

Table S4. Phylogenetic Distribution of Bacterial and Archaea Genomes in Groups A, B, C, D, and X
Table S4. Phylogenetic distribution of bacterial and archaea genomes in groups A, B, C, D, and X. Group A a: Total number of genomes in the taxon b: Number of group A genomes in the taxon c: Percentage of group A genomes in the taxon a b c cellular organisms 5007 2974 59.4 |__ Bacteria 4769 2935 61.5 | |__ Proteobacteria 1854 1570 84.7 | | |__ Gammaproteobacteria 711 631 88.7 | | | |__ Enterobacterales 112 97 86.6 | | | | |__ Enterobacteriaceae 41 32 78.0 | | | | | |__ unclassified Enterobacteriaceae 13 7 53.8 | | | | |__ Erwiniaceae 30 28 93.3 | | | | | |__ Erwinia 10 10 100.0 | | | | | |__ Buchnera 8 8 100.0 | | | | | | |__ Buchnera aphidicola 8 8 100.0 | | | | | |__ Pantoea 8 8 100.0 | | | | |__ Yersiniaceae 14 14 100.0 | | | | | |__ Serratia 8 8 100.0 | | | | |__ Morganellaceae 13 10 76.9 | | | | |__ Pectobacteriaceae 8 8 100.0 | | | |__ Alteromonadales 94 94 100.0 | | | | |__ Alteromonadaceae 34 34 100.0 | | | | | |__ Marinobacter 12 12 100.0 | | | | |__ Shewanellaceae 17 17 100.0 | | | | | |__ Shewanella 17 17 100.0 | | | | |__ Pseudoalteromonadaceae 16 16 100.0 | | | | | |__ Pseudoalteromonas 15 15 100.0 | | | | |__ Idiomarinaceae 9 9 100.0 | | | | | |__ Idiomarina 9 9 100.0 | | | | |__ Colwelliaceae 6 6 100.0 | | | |__ Pseudomonadales 81 81 100.0 | | | | |__ Moraxellaceae 41 41 100.0 | | | | | |__ Acinetobacter 25 25 100.0 | | | | | |__ Psychrobacter 8 8 100.0 | | | | | |__ Moraxella 6 6 100.0 | | | | |__ Pseudomonadaceae 40 40 100.0 | | | | | |__ Pseudomonas 38 38 100.0 | | | |__ Oceanospirillales 73 72 98.6 | | | | |__ Oceanospirillaceae -

Halanaerobaculum Jean Luc Cayol
Halanaerobaculum Jean Luc Cayol To cite this version: Jean Luc Cayol. Halanaerobaculum. Bergey’s Manual of Systematics of Archaea and Bacteria (BMSAB), Hoboken, New Jersey : Wiley, [2015]-, 2019, 10.1002/9781118960608.gbm01725. hal- 02883868 HAL Id: hal-02883868 https://hal.archives-ouvertes.fr/hal-02883868 Submitted on 29 Jun 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 gbm01725 2 3 Halanaerobaculum 4 5 Hedi, Fardeau, Sadfi, Boudabous, Ollivier and Cayol 2009, 317 VP 6 7 Jean-Luc Cayol 8 9 Aix Marseille Université, IRD, Université de Toulon, CNRS, Mediterranean Institute of 10 Oceanography (MIO), UM 110, 13288 Marseille cedex 9, France 11 12 Hal.an.ae.ro.ba’cu.lum. Gr. masc. n. hals, salt; Gr. pref. an, not; Gr. masc. or fem. n. aer, air; 13 L. neut. n. baculum, stick; N.L. neut. n. Halanaerobaculum, salt stick not living in air. 14 15 Abstract 16 The genus Halanaerobaculum comprises one species with validly published name, 17 Halanaerobaculum tunisiense, which was isolated from hypersaline surface sediments of El- 18 Djerid Chott, Tunisia. The genus is halophilic, strict anaerobic and chemoorganotrophic. The 19 genus Halanaerobaculum belongs to the family Halobacteroidaceae; order Halanaerobiales; 20 class Clostridia.