Compile.Xlsx
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Patterns, Processes and Models of Microbial Recovery in a Chronosequence Following Reforestation of Reclaimed Mine Soils
PATTERNS, PROCESSES AND MODELS OF MICROBIAL RECOVERY IN A CHRONOSEQUENCE FOLLOWING REFORESTATION OF RECLAIMED MINE SOILS Shan Sun Dissertation submitted to the faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Doctor of Philosophy In Crop and Soil Environmental Sciences Brian Badgley, Chair Song Li Brian Strahm Michael Strickland Carl Zipper Virginia Polytechnic Institute and State University Blacksburg, Virginia July 2017 Keywords: soil microorganism, recovery, chronosequence, reclaimed mine soils, amplicon sequencing, shotgun metagenomics, bacterial co-occurrence groups, artificial neural network PATTERNS, PROCESSES AND MODELS OF MICROBIAL RECOVERY IN A CHRONOSEQUENCE FOLLOWING REFORESTATION OF RECLAIMED MINE SOILS Shan Sun Abstract Soil microbial communities mediate important ecological processes and play essential roles in biogeochemical cycling. Ecosystem disturbances such as surface mining significantly alter soil microbial communities, which could lead to changes or impairment of ecosystem functions. Reforestation procedures were designed to accelerate the reestablishment of plant community and the recovery of the forest ecosystem after reclamation. However, the microbial recovery during reforestation has not been well studied even though this information is essential for evaluating ecosystem restoration success. In addition, the similar starting conditions of mining sites of different ages facilitate a chronosequence approach for studying decades-long microbial community change, which could help generalize theories about ecosystem succession. In this study, the recovery of microbial communities in a chronosequence of reclaimed mine sites spanning 30 years post reforestation along with unmined reference sites was analyzed using next-generation sequencing to characterize soil-microbial abundance, richness, taxonomic composition, interaction patterns and functional genes. -

Metaproteogenomic Insights Beyond Bacterial Response to Naphthalene
ORIGINAL ARTICLE ISME Journal – Original article Metaproteogenomic insights beyond bacterial response to 5 naphthalene exposure and bio-stimulation María-Eugenia Guazzaroni, Florian-Alexander Herbst, Iván Lores, Javier Tamames, Ana Isabel Peláez, Nieves López-Cortés, María Alcaide, Mercedes V. del Pozo, José María Vieites, Martin von Bergen, José Luis R. Gallego, Rafael Bargiela, Arantxa López-López, Dietmar H. Pieper, Ramón Rosselló-Móra, Jesús Sánchez, Jana Seifert and Manuel Ferrer 10 Supporting Online Material includes Text (Supporting Materials and Methods) Tables S1 to S9 Figures S1 to S7 1 SUPPORTING TEXT Supporting Materials and Methods Soil characterisation Soil pH was measured in a suspension of soil and water (1:2.5) with a glass electrode, and 5 electrical conductivity was measured in the same extract (diluted 1:5). Primary soil characteristics were determined using standard techniques, such as dichromate oxidation (organic matter content), the Kjeldahl method (nitrogen content), the Olsen method (phosphorus content) and a Bernard calcimeter (carbonate content). The Bouyoucos Densimetry method was used to establish textural data. Exchangeable cations (Ca, Mg, K and 10 Na) extracted with 1 M NH 4Cl and exchangeable aluminium extracted with 1 M KCl were determined using atomic absorption/emission spectrophotometry with an AA200 PerkinElmer analyser. The effective cation exchange capacity (ECEC) was calculated as the sum of the values of the last two measurements (sum of the exchangeable cations and the exchangeable Al). Analyses were performed immediately after sampling. 15 Hydrocarbon analysis Extraction (5 g of sample N and Nbs) was performed with dichloromethane:acetone (1:1) using a Soxtherm extraction apparatus (Gerhardt GmbH & Co. -

Microbial Diversity of Non-Flooded High Temperature Petroleum Reservoir in South of Iran
Archive of SID Biological Journal of Microorganism th 8 Year, Vol. 8, No. 32, Winter 2020 Received: November 18, 2018/ Accepted: May 21, 2019. Page: 15-231- 8 Microbial Diversity of Non-flooded High Temperature Petroleum Reservoir in South of Iran Mohsen Pournia Department of Microbiology, Shiraz Branch, Islamic Azad University, Shiraz, Iran, [email protected] Nima Bahador * Department of Microbiology, Shiraz Branch, Islamic Azad University, Shiraz, Iran, [email protected] Meisam Tabatabaei Biofuel Research Team (BRTeam), Karaj, Iran, [email protected] Reza Azarbayjani Molecular bank, Iranian Biological Resource Center, ACECR, Karaj, Iran, [email protected] Ghassem Hosseni Salekdeh Department of Biology, Agricultural Biotechnology Research Institute, Karaj, Iran, [email protected] Abstract Introduction: Although bacteria and archaea are able to grow and adapted to the petrol reservoirs during several years, there are no results from microbial diversity of oilfields with high temperature in Iran. Hence, the present study tried to identify microbial community in non-water flooding Zeilaei (ZZ) oil reservoir. Materials and methods: In this study, for the first time, non-water flooded high temperature Zeilaei oilfield was analyzed for its microbial community based on next generation sequencing of 16S rRNA genes. Results: The results obtained from this study indicated that the most abundant bacterial community belonged to phylum of Firmicutes (Bacilli ) and Thermotoga, while other phyla (Proteobacteria , Actinobacteria and Synergistetes ) were much less abundant. Bacillus subtilis , B. licheniformis , Petrotoga mobilis , P. miotherma, Fervidobacterium pennivorans , and Thermotoga subterranea were observed with high frequency. In addition, the most abundant archaea were Methanothermobacter thermautotrophicus . Discussion and conclusion: Although there are many reports on the microbial community of oil filed reservoirs, this is the first report of large quantities of Bacillus spp. -

Identification of Functional Lsrb-Like Autoinducer-2 Receptors
Swarthmore College Works Chemistry & Biochemistry Faculty Works Chemistry & Biochemistry 11-15-2009 Identification Of unctionalF LsrB-Like Autoinducer-2 Receptors C. S. Pereira Anna Katherine De Regt , '09 P. H. Brito Stephen T. Miller Swarthmore College, [email protected] K. B. Xavier Follow this and additional works at: https://works.swarthmore.edu/fac-chemistry Part of the Biochemistry Commons Let us know how access to these works benefits ouy Recommended Citation C. S. Pereira; Anna Katherine De Regt , '09; P. H. Brito; Stephen T. Miller; and K. B. Xavier. (2009). "Identification Of unctionalF LsrB-Like Autoinducer-2 Receptors". Journal Of Bacteriology. Volume 191, Issue 22. 6975-6987. DOI: 10.1128/JB.00976-09 https://works.swarthmore.edu/fac-chemistry/52 This work is brought to you for free by Swarthmore College Libraries' Works. It has been accepted for inclusion in Chemistry & Biochemistry Faculty Works by an authorized administrator of Works. For more information, please contact [email protected]. Identification of Functional LsrB-Like Autoinducer-2 Receptors Catarina S. Pereira, Anna K. de Regt, Patrícia H. Brito, Stephen T. Miller and Karina B. Xavier J. Bacteriol. 2009, 191(22):6975. DOI: 10.1128/JB.00976-09. Published Ahead of Print 11 September 2009. Downloaded from Updated information and services can be found at: http://jb.asm.org/content/191/22/6975 http://jb.asm.org/ These include: SUPPLEMENTAL MATERIAL Supplemental material REFERENCES This article cites 65 articles, 29 of which can be accessed free on September 10, 2014 by SWARTHMORE COLLEGE at: http://jb.asm.org/content/191/22/6975#ref-list-1 CONTENT ALERTS Receive: RSS Feeds, eTOCs, free email alerts (when new articles cite this article), more» Information about commercial reprint orders: http://journals.asm.org/site/misc/reprints.xhtml To subscribe to to another ASM Journal go to: http://journals.asm.org/site/subscriptions/ JOURNAL OF BACTERIOLOGY, Nov. -

Deciphering a Marine Bone Degrading Microbiome Reveals a Complex Community Effort
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.13.093005; this version posted November 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Deciphering a marine bone degrading microbiome reveals a complex community effort 2 3 Erik Borcherta,#, Antonio García-Moyanob, Sergio Sanchez-Carrilloc, Thomas G. Dahlgrenb,d, 4 Beate M. Slabya, Gro Elin Kjæreng Bjergab, Manuel Ferrerc, Sören Franzenburge and Ute 5 Hentschela,f 6 7 aGEOMAR Helmholtz Centre for Ocean Research Kiel, RD3 Research Unit Marine Symbioses, 8 Kiel, Germany 9 bNORCE Norwegian Research Centre, Bergen, Norway 10 cCSIC, Institute of Catalysis, Madrid, Spain 11 dDepartment of Marine Sciences, University of Gothenburg, Gothenburg, Sweden 12 eIKMB, Institute of Clinical Molecular Biology, University of Kiel, Kiel, Germany 13 fChristian-Albrechts University of Kiel, Kiel, Germany 14 15 Running Head: Marine bone degrading microbiome 16 #Address correspondence to Erik Borchert, [email protected] 17 Abstract word count: 229 18 Text word count: 4908 (excluding Abstract, Importance, Materials and Methods) 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.05.13.093005; this version posted November 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 19 Abstract 20 The marine bone biome is a complex assemblage of macro- and microorganisms, however the 21 enzymatic repertoire to access bone-derived nutrients remains unknown. -

Desulfuribacillus Alkaliarsenatis Gen. Nov. Sp. Nov., a Deep-Lineage
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Extremophiles (2012) 16:597–605 DOI 10.1007/s00792-012-0459-7 ORIGINAL PAPER Desulfuribacillus alkaliarsenatis gen. nov. sp. nov., a deep-lineage, obligately anaerobic, dissimilatory sulfur and arsenate-reducing, haloalkaliphilic representative of the order Bacillales from soda lakes D. Y. Sorokin • T. P. Tourova • M. V. Sukhacheva • G. Muyzer Received: 10 February 2012 / Accepted: 3 May 2012 / Published online: 24 May 2012 Ó The Author(s) 2012. This article is published with open access at Springerlink.com Abstract An anaerobic enrichment culture inoculated possible within a pH range from 9 to 10.5 (optimum at pH with a sample of sediments from soda lakes of the Kulunda 10) and a salt concentration at pH 10 from 0.2 to 2 M total Steppe with elemental sulfur as electron acceptor and for- Na? (optimum at 0.6 M). According to the phylogenetic mate as electron donor at pH 10 and moderate salinity analysis, strain AHT28 represents a deep independent inoculated with sediments from soda lakes in Kulunda lineage within the order Bacillales with a maximum of Steppe (Altai, Russia) resulted in the domination of a 90 % 16S rRNA gene similarity to its closest cultured Gram-positive, spore-forming bacterium strain AHT28. representatives. On the basis of its distinct phenotype and The isolate is an obligate anaerobe capable of respiratory phylogeny, the novel haloalkaliphilic anaerobe is suggested growth using elemental sulfur, thiosulfate (incomplete as a new genus and species, Desulfuribacillus alkaliar- T T reduction) and arsenate as electron acceptor with H2, for- senatis (type strain AHT28 = DSM24608 = UNIQEM mate, pyruvate and lactate as electron donor. -

Bacterial Diversity in the Surface Sediments of the Hypoxic Zone Near
ORIGINAL RESEARCH Bacterial diversity in the surface sediments of the hypoxic zone near the Changjiang Estuary and in the East China Sea Qi Ye, Ying Wu, Zhuoyi Zhu, Xiaona Wang, Zhongqiao Li & Jing Zhang State Key Laboratory of Estuarine and Coastal Research, East China Normal University, Shanghai 200062, China Keywords Abstract Bacteria, Changjiang Estuary, hypoxia, Miseq Illumina sequencing, sediment Changjiang (Yangtze River) Estuary has experienced severe hypoxia since the 1950s. In order to investigate potential ecological functions of key microorgan- Correspondence isms in relation to hypoxia, we performed 16S rRNA-based Illumina Miseq Qi Ye, East China Normal University, State Key sequencing to explore the bacterial diversity in the surface sediments of the Laboratory of Estuarine and Coastal Research, hypoxic zone near the Changjiang Estuary and in the East China Sea (ECS). 3663 North Zhongshan Road, SKLEC Building, Room 419, Shanghai 200062, China. The results showed that numerous Proteobacteria-affiliated sequences in the sedi- Tel: 86-021-52124974; ments of the inner continental shelf were related to both sulfate-reducing and Fax: 86-021- 62546441; sulfur-oxidizing bacteria, suggesting an active sulfur cycle in this area. Many E-mail: [email protected] sequences retrieved from the hypoxic zone were also related to Planctomycetes from two marine upwelling systems, which may be involved in the initial break- Funding Information down of sulfated heteropolysaccharides. Bacteroidetes, which is expected to degrade This study was funded by the Shanghai Pujiang high-molecular-weight organic matter, was abundant in all the studied stations Talent Program (12PJ1403100), the National except for station A8, which was the deepest and possessed the largest grain Natural Science Foundation of China (41276081), the Key Project of Chinese size. -

Successful Drug Discovery Informed by Actinobacterial Systematics
Successful Drug Discovery Informed by Actinobacterial Systematics Verrucosispora HPLC-DAD analysis of culture filtrate Structures of Abyssomicins Biological activity T DAD1, 7.382 (196 mAU,Up2) of 002-0101.D V. maris AB-18-032 mAU CH3 CH3 T extract H3C H3C Antibacterial activity (MIC): S. leeuwenhoekii C34 maris AB-18-032 175 mAU DAD1 A, Sig=210,10 150 C DAD1 B, Sig=230,10 O O DAD1 C, Sig=260,20 125 7 7 500 Rt 7.4 min DAD1 D, Sig=280,20 O O O O Growth inhibition of Gram-positive bacteria DAD1 , Sig=310,20 100 Abyssomicins DAD1 F, Sig=360,40 C 75 DAD1 G, Sig=435,40 Staphylococcus aureus (MRSA) 4 µg/ml DAD1 H, Sig=500,40 50 400 O O 25 O O Staphylococcus aureus (iVRSA) 13 µg/ml 0 CH CH3 300 400 500 nm 3 DAD1, 7.446 (300 mAU,Dn1) of 002-0101.D 300 mAU Mode of action: C HO atrop-C HO 250 atrop-C CH3 CH3 CH3 CH3 200 H C H C H C inhibitior of pABA biosynthesis 200 Rt 7.5 min H3C 3 3 3 Proximicin A Proximicin 150 HO O HO O O O O O O O O O A 100 O covalent binding to Cys263 of PabB 100 N 50 O O HO O O Sea of Japan B O O N O O (4-amino-4-deoxychorismate synthase) by 0 CH CH3 CH3 CH3 3 300 400 500 nm HO HO HO HO Michael addition -289 m 0 B D G H 2 4 6 8 10 12 14 16 min Newcastle Michael Goodfellow, School of Biology, University Newcastle University, Newcastle upon Tyne Atacama Desert In This Talk I will Consider: • Actinobacteria as a key group in the search for new therapeutic drugs. -

Microbiome Exploration of Deep-Sea Carnivorous Cladorhizidae Sponges
Microbiome exploration of deep-sea carnivorous Cladorhizidae sponges by Joost Theo Petra Verhoeven A Thesis submitted to the School of Graduate Studies in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Biology Memorial University of Newfoundland March 2019 St. John’s, Newfoundland and Labrador ABSTRACT Members of the sponge family Cladorhizidae are unique in having replaced the typical filter-feeding strategy of sponges by a predatory lifestyle, capturing and digesting small prey. These carnivorous sponges are found in many different environments, but are particularly abundant in deep waters, where they constitute a substantial component of the benthos. Sponges are known to host a wide range of microbial associates (microbiome) important for host health, but the extent of the microbiome in carnivorous sponges has never been extensively investigated and their importance is poorly understood. In this thesis, the microbiome of two deep-sea carnivorous sponge species (Chondrocladia grandis and Cladorhiza oxeata) is investigated for the first time, leveraging recent advances in high-throughput sequencing and through custom developed bioinformatic and molecular methods. Microbiome analyses showed that the carnivorous sponges co-occur with microorganisms and large differences in the composition and type of associations were observed between sponge species. Tissues of C. grandis hosted diverse bacterial communities, similar in composition between individuals, in stark contrast to C. oxeata where low microbial diversity was found with a high host-to-host variability. In C. grandis the microbiome was not homogeneous throughout the host tissue, and significant shifts occured within community members across anatomical regions, with the enrichment of specific bacterial taxa in particular anatomical niches, indicating a potential symbiotic role of such taxa within processes like prey digestion and chemolithoautotrophy. -
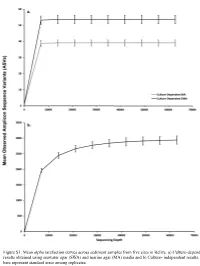
Figure S1. Mean Alpha Rarefaction Curves Across Sediment Samples from Five Sites in Belize
Figure S1. Mean alpha rarefaction curves across sediment samples from five sites in Belize. a) Culture-dependent results obtained using seawater agar (SWA) and marine agar (MA) media and b) Culture- independent results. Error bars represent standard error among replicates. a. 200 150 100 Faith’s PD Faith’s 50 0 Culturedependent MA Culturedependent SWA Cultureindependent Method b. 0.8 0.6 Pielou’s Evenness 0.4 Culturedependent MA Culturedependent SWA Cultureindependent Method Figure S2. Alpha diversity boxplots of marine sediment microbial communities from Carrie Bow Cay, Belize in culture-dependent and culture-independent samples determined using a) Faith’s Phylogenetic Diversity Index and b) Pielou’s Evenness. Culture-dependent methods include the use of marine agar medium (MA) and seawater agar medium (SWA. Data points are overlayed on the boxplot to show variation. a. 100% Proteobacteria Nitrospinae Bacteroidetes Fibrobacteres Planctomycetes Entotheonellaeota 90% Cyanobacteria Marinimicrobia (SAR406 clade) Firmicutes Deinococcus-Thermus Chloroflexi Margulisbacteria [A] Thaumarchaeota Unassigned 80% Acidobacteria Archaea UA Verrucomicrobia Acetothermia Actinobacteria Elusimicrobia 70% [A] Nanoarchaeaeota Tenericutes Kiritimatiellaeota TA06 Latescibacteria WS2 60% Spirochaetes Dependentiae Patescibacteria LCP-89 Gemmatimonadetes FCPU426 Omnitrophicaeota WPS-2 50% Fusobacteria CK-2C2-2 Bacteria UA Armatimonadetes [A] Euryarchaeota [A] Altiarchaeota Relative Percent 40% Calditrichaeota Poribacteria Lentisphaerae Cloacimonetes [A] Crenarchaeota -

Yu-Chen Ling and John W. Moreau
Microbial Distribution and Activity in a Coastal Acid Sulfate Soil System Introduction: Bioremediation in Yu-Chen Ling and John W. Moreau coastal acid sulfate soil systems Method A Coastal acid sulfate soil (CASS) systems were School of Earth Sciences, University of Melbourne, Melbourne, VIC 3010, Australia formed when people drained the coastal area Microbial distribution controlled by environmental parameters Microbial activity showed two patterns exposing the soil to the air. Drainage makes iron Microbial structures can be grouped into three zones based on the highest similarity between samples (Fig. 4). Abundant populations, such as Deltaproteobacteria, kept constant activity across tidal cycling, whereas rare sulfides oxidize and release acidity to the These three zones were consistent with their geological background (Fig. 5). Zone 1: Organic horizon, had the populations changed activity response to environmental variations. Activity = cDNA/DNA environment, low pH pore water further dissolved lowest pH value. Zone 2: surface tidal zone, was influenced the most by tidal activity. Zone 3: Sulfuric zone, Abundant populations: the heavy metals. The acidity and toxic metals then Method A Deltaproteobacteria Deltaproteobacteria this area got neutralized the most. contaminate coastal and nearby ecosystems and Method B 1.5 cause environmental problems, such as fish kills, 1.5 decreased rice yields, release of greenhouse gases, Chloroflexi and construction damage. In Australia, there is Gammaproteobacteria Gammaproteobacteria about a $10 billion “legacy” from acid sulfate soils, Chloroflexi even though Australia is only occupied by around 1.0 1.0 Cyanobacteria,@ Acidobacteria Acidobacteria Alphaproteobacteria 18% of the global acid sulfate soils. Chloroplast Zetaproteobacteria Rare populations: Alphaproteobacteria Method A log(RNA(%)+1) Zetaproteobacteria log(RNA(%)+1) Method C Method B 0.5 0.5 Cyanobacteria,@ Bacteroidetes Chloroplast Firmicutes Firmicutes Bacteroidetes Planctomycetes Planctomycetes Ac8nobacteria Fig. -

Dissertation Implementing Organic Amendments To
DISSERTATION IMPLEMENTING ORGANIC AMENDMENTS TO ENHANCE MAIZE YIELD, SOIL MOISTURE, AND MICROBIAL NUTRIENT CYCLING IN TEMPERATE AGRICULTURE Submitted by Erika J. Foster Graduate Degree Program in Ecology In partial fulfillment of the requirements For the Degree of Doctor of Philosophy Colorado State University Fort Collins, Colorado Summer 2018 Doctoral Committee: Advisor: M. Francesca Cotrufo Louise Comas Charles Rhoades Matthew D. Wallenstein Copyright by Erika J. Foster 2018 All Rights Reserved i ABSTRACT IMPLEMENTING ORGANIC AMENDMENTS TO ENHANCE MAIZE YIELD, SOIL MOISTURE, AND MICROBIAL NUTRIENT CYCLING IN TEMPERATE AGRICULTURE To sustain agricultural production into the future, management should enhance natural biogeochemical cycling within the soil. Strategies to increase yield while reducing chemical fertilizer inputs and irrigation require robust research and development before widespread implementation. Current innovations in crop production use amendments such as manure and biochar charcoal to increase soil organic matter and improve soil structure, water, and nutrient content. Organic amendments also provide substrate and habitat for soil microorganisms that can play a key role cycling nutrients, improving nutrient availability for crops. Additional plant growth promoting bacteria can be incorporated into the soil as inocula to enhance soil nutrient cycling through mechanisms like phosphorus solubilization. Since microbial inoculation is highly effective under drought conditions, this technique pairs well in agricultural systems using limited irrigation to save water, particularly in semi-arid regions where climate change and population growth exacerbate water scarcity. The research in this dissertation examines synergistic techniques to reduce irrigation inputs, while building soil organic matter, and promoting natural microbial function to increase crop available nutrients. The research was conducted on conventional irrigated maize systems at the Agricultural Research Development and Education Center north of Fort Collins, CO.