(NORMALIZED to the Genus Level): SRB/SOB
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Updating the Taxonomic Toolbox: Classification of Alteromonas Spp
1 Updating the taxonomic toolbox: classification of Alteromonas spp. 2 using Multilocus Phylogenetic Analysis and MALDI-TOF Mass 3 Spectrometry a a a 4 Hooi Jun Ng , Hayden K. Webb , Russell J. Crawford , François a b b c 5 Malherbe , Henry Butt , Rachel Knight , Valery V. Mikhailov and a, 6 Elena P. Ivanova * 7 aFaculty of Life and Social Sciences, Swinburne University of Technology, 8 PO Box 218, Hawthorn, Vic 3122, Australia 9 bBioscreen, Bio21 Institute, The University of Melbourne, Vic 3010, Australia 10 cG.B. Elyakov Pacific Institute of Bioorganic Chemistry, Far Eastern Branch, Russian 11 Academy of Sciences, Vladivostok 690022, Russian Federation 12 13 *Corresponding author: Tel: +61-3-9214-5137. Fax: +61-3-9214-5050. 14 E-mail: [email protected] 15 16 Abstract 17 Bacteria of the genus Alteromonas are Gram-negative, strictly aerobic, motile, 18 heterotrophic marine bacteria, known for their versatile metabolic activities. 19 Identification and classification of novel species belonging to the genus Alteromonas 20 generally involves DNA-DNA hybridization (DDH) as distinct species often fail to be 1 21 resolved at the 97% threshold value of the 16S rRNA gene sequence similarity. In this 22 study, the applicability of Multilocus Phylogenetic Analysis (MLPA) and Matrix- 23 Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF 24 MS) for the differentiation of Alteromonas species has been evaluated. Phylogenetic 25 analysis incorporating five house-keeping genes (dnaK, sucC, rpoB, gyrB, and rpoD) 26 revealed a threshold value of 98.9% that could be considered as the species cut-off 27 value for the delineation of Alteromonas spp. -

Supplementary Materials
SUPPLEMENTARY MATERIALS Table S1. Chemical characteristics of the two digestate forms (SD and WD). Values quoted are expressed as % of air-dry digestate (means followed by standard error in brackets). References for the employed methods used for determination of each chemical characteristic is also reported. SD WD Reference Org C % 44.4 (0.33) 1.1 (0.01) [81] Tot N % 1.4 (0.01) 0.4 (0.01) [82] C/N 31.4 (0.17) 3.1 (0.04) NH4-N % n.d 0.2 (0.00) [83] K % 1.7 (0.00) n.d. [84] P % 0.9 (0.01) n.d. [84] S % 0.23 (0.02) n.d. [85] SD = solid digestate; WD = whole digestate. Table S2. Soil physical and chemical characteristics at the beginning of trial (t0) (means from 9 observations followed by standard errors in brackets). Clay (%) 41.9 (1.22) Silt (%) 47.8 (2.13) Moisture (%) 24.46 (1.24) Bulk density (g cm-3) 1.39 (0.04) pH 8.3 (0) CaCO3 (%) 11.4 (0.7) TOC (g kg-1) 12.8 (0.3) TN (g kg-1) 1.4 (0) C/N 9.4 (0.3) CEC (cmol(+) kg-1) 21.0 (0.7) Exchangeable Bases (mg kg-1) K 278.7 (8.0) Na 22.0 (3.0) Mg 201.7 (25.6) Ca 3718.6 (176.3) Available Microelements (mg kg-1) Cu 28.0 (5.0) Zn 1.7 (0.2) Fe 15.4 (0.5) Mn 16.3 (0.5) TOC = total organic C; TN = total N; CEC = cation exchange capacity Table S3. -

UNIVERSITY of CALIFORNIA, SAN DIEGO Indicators of Iron
UNIVERSITY OF CALIFORNIA, SAN DIEGO Indicators of Iron Metabolism in Marine Microbial Genomes and Ecosystems A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Oceanography by Shane Lahman Hogle Committee in charge: Katherine Barbeau, Chair Eric Allen Bianca Brahamsha Christopher Dupont Brian Palenik Kit Pogliano 2016 Copyright Shane Lahman Hogle, 2016 All rights reserved . The Dissertation of Shane Lahman Hogle is approved, and it is acceptable in quality and form for publication on microfilm and electronically: Chair University of California, San Diego 2016 iii DEDICATION Mom, Dad, Joel, and Marie thank you for everything iv TABLE OF CONTENTS Signature Page ................................................................................................................... iii Dedication .......................................................................................................................... iv Table of Contents .................................................................................................................v List of Figures ................................................................................................................... vii List of Tables ..................................................................................................................... ix Acknowledgements ..............................................................................................................x Vita .................................................................................................................................. -

1 Supplementary Information Ugly Ducklings – the Dark Side of Plastic
Supplementary Information Ugly ducklings – The dark side of plastic materials in contact with potable water Lisa Neu1,2, Carola Bänziger1, Caitlin R. Proctor1,2, Ya Zhang3, Wen-Tso Liu3, Frederik Hammes1,* 1 Eawag, Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, Switzerland 2 Department of Environmental Systems Science, Institute of Biogeochemistry and Pollutant Dynamics, ETH Zürich, Zürich, Switzerland 3 Department of Civil and Environmental Engineering, University of Illinois at Urbana-Champaign, USA Table of contents Table S1 Exemplary online blog entries on biofouling in bath toys Figure S1 Images of all examined bath toys Figure S2 Additional images of bath toy biofilms by OCT Figure S3 Additional images on biofilm composition by SEM Figure S4 Number of bacteria and proportion of intact cells in bath toy biofilms Table S2 Classification of shared OTUs between bath toys Table S3 Shared and ‘core’ communities in bath toys from single households Table S4 Richness and diversity Figure S5 Classification of abundant OTUs in real bath toy biofilms Table S5 Comparison of most abundant OTUs in control bath toy biofilms Figure S6 Fungal community composition in bath toy biofilms Table S6 Conventional plating results for indicator bacteria and groups Table S7 Bioavailability of migrating carbon from control bath toys’ material Water chemistry Method and results (Table S8) Table S9 Settings for Amplification PCR and Index PCR reactions 1 Table S1: Exemplary online blog entries on biofouling inside bath toys Issue - What is the slime? Link Rub-a-dub-dub, https://www.babble.com/baby/whats-in-the-tub/ what’s in the tub? What’s the black stuff http://blogs.babycenter.com/momstories/whats-the-black- in your squeeze toys? stuff-in-your-squeeze-toys/ Friday Find: NBC’s http://www.bebravekeepgoing.com/2010/03/friday-find-nbcs- Today Show segment: today-show-segment-do.html Do bath toys carry germs? Yuck. -
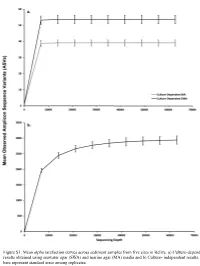
Figure S1. Mean Alpha Rarefaction Curves Across Sediment Samples from Five Sites in Belize
Figure S1. Mean alpha rarefaction curves across sediment samples from five sites in Belize. a) Culture-dependent results obtained using seawater agar (SWA) and marine agar (MA) media and b) Culture- independent results. Error bars represent standard error among replicates. a. 200 150 100 Faith’s PD Faith’s 50 0 Culturedependent MA Culturedependent SWA Cultureindependent Method b. 0.8 0.6 Pielou’s Evenness 0.4 Culturedependent MA Culturedependent SWA Cultureindependent Method Figure S2. Alpha diversity boxplots of marine sediment microbial communities from Carrie Bow Cay, Belize in culture-dependent and culture-independent samples determined using a) Faith’s Phylogenetic Diversity Index and b) Pielou’s Evenness. Culture-dependent methods include the use of marine agar medium (MA) and seawater agar medium (SWA. Data points are overlayed on the boxplot to show variation. a. 100% Proteobacteria Nitrospinae Bacteroidetes Fibrobacteres Planctomycetes Entotheonellaeota 90% Cyanobacteria Marinimicrobia (SAR406 clade) Firmicutes Deinococcus-Thermus Chloroflexi Margulisbacteria [A] Thaumarchaeota Unassigned 80% Acidobacteria Archaea UA Verrucomicrobia Acetothermia Actinobacteria Elusimicrobia 70% [A] Nanoarchaeaeota Tenericutes Kiritimatiellaeota TA06 Latescibacteria WS2 60% Spirochaetes Dependentiae Patescibacteria LCP-89 Gemmatimonadetes FCPU426 Omnitrophicaeota WPS-2 50% Fusobacteria CK-2C2-2 Bacteria UA Armatimonadetes [A] Euryarchaeota [A] Altiarchaeota Relative Percent 40% Calditrichaeota Poribacteria Lentisphaerae Cloacimonetes [A] Crenarchaeota -

Redalyc.Shallow-Water Hydrothermal Vents in the Azores (Portugal)
Revista de Gestão Costeira Integrada - Journal of Integrated Coastal Zone Management E-ISSN: 1646-8872 [email protected] Associação Portuguesa dos Recursos Hídricos Portugal Couto, Ruben P.; Rodriguesa, Armindo S.; Neto, Ana I. Shallow-water hydrothermal vents in the Azores (Portugal) Revista de Gestão Costeira Integrada - Journal of Integrated Coastal Zone Management, vol. 15, núm. 4, 2015, pp. 495-505 Associação Portuguesa dos Recursos Hídricos Lisboa, Portugal Available in: http://www.redalyc.org/articulo.oa?id=388343047005 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Revista de Gestão Costeira Integrada / Journal of Integrated Coastal Zone Management, 15(4):495-505 (2015) http://www.aprh.pt/rgci/pdf/rgci-584_Couto.pdf | DOI: 10.5894/rgci584 Shallow-water hydrothermal vents in the Azores (Portugal)* @, Ruben P. Couto@, a, b; Armindo S. Rodriguesa, c; Ana I. Netoa, d ABSTRACT The impact of global warming has been a major issue in recent years and will continue increasing in the future. Knowledge about the effects of ocean acidification on marine organisms and communities is crucial to efficient management. Island envi- ronments are particularly sensitive to externally induced changes and highly dependent on their coastal areas. This study summarises the published information on shallow-water hydrothermal vents of the Azores. These environments were reported to exhibit high metal concentration and acidified seawater due to the diffusion of acidic volcanic gases (mainly CO2) and a considerable temperature range. -

Supplementary Information for Microbial Electrochemical Systems Outperform Fixed-Bed Biofilters for Cleaning-Up Urban Wastewater
Electronic Supplementary Material (ESI) for Environmental Science: Water Research & Technology. This journal is © The Royal Society of Chemistry 2016 Supplementary information for Microbial Electrochemical Systems outperform fixed-bed biofilters for cleaning-up urban wastewater AUTHORS: Arantxa Aguirre-Sierraa, Tristano Bacchetti De Gregorisb, Antonio Berná, Juan José Salasc, Carlos Aragónc, Abraham Esteve-Núñezab* Fig.1S Total nitrogen (A), ammonia (B) and nitrate (C) influent and effluent average values of the coke and the gravel biofilters. Error bars represent 95% confidence interval. Fig. 2S Influent and effluent COD (A) and BOD5 (B) average values of the hybrid biofilter and the hybrid polarized biofilter. Error bars represent 95% confidence interval. Fig. 3S Redox potential measured in the coke and the gravel biofilters Fig. 4S Rarefaction curves calculated for each sample based on the OTU computations. Fig. 5S Correspondence analysis biplot of classes’ distribution from pyrosequencing analysis. Fig. 6S. Relative abundance of classes of the category ‘other’ at class level. Table 1S Influent pre-treated wastewater and effluents characteristics. Averages ± SD HRT (d) 4.0 3.4 1.7 0.8 0.5 Influent COD (mg L-1) 246 ± 114 330 ± 107 457 ± 92 318 ± 143 393 ± 101 -1 BOD5 (mg L ) 136 ± 86 235 ± 36 268 ± 81 176 ± 127 213 ± 112 TN (mg L-1) 45.0 ± 17.4 60.6 ± 7.5 57.7 ± 3.9 43.7 ± 16.5 54.8 ± 10.1 -1 NH4-N (mg L ) 32.7 ± 18.7 51.6 ± 6.5 49.0 ± 2.3 36.6 ± 15.9 47.0 ± 8.8 -1 NO3-N (mg L ) 2.3 ± 3.6 1.0 ± 1.6 0.8 ± 0.6 1.5 ± 2.0 0.9 ± 0.6 TP (mg -

Bacterial Profiles and Antibiogrants of the Bacteria Isolated of the Exposed Pulps of Dog and Cheetah Canine Teeth
Bacterial profiles and antibiogrants of the bacteria isolated of the exposed pulps of dog and cheetah canine teeth A dissertation submitted to the Faculty of Veterinary Science, University of Pretoria. In partial fulfillment of the requirements for the degree Master of Science (Veterinary Science) Promoter: Dr. Gerhard Steenkamp Co-promoter: Ms. Anna-Mari Bosman Department of Companion Animal Clinical Studies Faculty of Veterinary Science University of Pretoria Pretoria (January 2012) J.C. Almansa Ruiz © University of Pretoria Declaration I declare that the dissertation that I hereby submit for the Masters of Science degree in Veterinary Science at the University of Pretoria has not previously been submitted by me for degree purposes at any other university. J.C. Almansa Ruiz ii Dedications To one of the most amazing hunters of the African bush, the Cheetah, that has made me dream since I was a child, and the closest I had been to one, before starting this project was in National Geographic documentaries. I wish all of them a better future in which their habitat will be more respected. To all conservationists, especially to Carla Conradie and Dave Houghton, for spending their lives saving these animals which are suffering from the consequences of the encroachment of human beings into their territory. Some, such as George Adamson, the lion conservationist, even lost their lives in this mission. To the conservationist, Lawrence Anthony, for risking his life in a suicide mission to save the animals in the Baghdad Zoo, when the conflict in Iraq exploded. To my mother and father Jose Maria and Rosa. -
Albirhodobacter Marinus Gen. Nov., Sp. Nov., a Member of the Family Rhodobacteriaceae Isolated from Sea Shore Water of Visakhapatnam, India
Author version: Antonie van Leeuwenhoek, vol.103; 2013; 347-355 Albirhodobacter marinus gen. nov., sp. nov., a member of the family Rhodobacteriaceae isolated from sea shore water of Visakhapatnam, India Nupur1, Bhumika, Vidya1., Srinivas, T. N. R2,3, Anil Kumar, P1* 1Microbial Type Culture Collection and Gene bank, Institute of Microbial Technology (CSIR), Sector 39A, Chandigarh - 160 036, INDIA 2National Institute of Oceanography (CSIR), Regional centre, P B No. 1913, Dr. Salim Ali Road, Kochi - 682018 (Kerala), INDIA Present Address: 3National Institute of Oceanography (CSIR), Regional centre, 176, Lawsons Bay Colony, Visakhapatnam - 530 017 (Andhra Pradesh), INDIA Address for correspondence* Dr. P. Anil Kumar Microbial Type Culture Collection and Gene bank Institute of Microbial Technology, Sector 39A, Chandigarh - 160 036, INDIA Email: [email protected] Phone: +91-172-6665170 1 Abstract A novel marine, Gram-negative, rod-shaped bacterium, designated strain N9T, was isolated from a water sample of the sea shore at Visakhapatnam, Andhra Pradesh (India). Strain N9T was found to be positive for oxidase and catalase activities. The fatty acids were found to be dominated by C16:0, C18:1 ω7c and summed in feature 3 (C16:1 ω7c and/or C16:1 ω6c). Strain N9T was determined to contain Q-10 as the major respiratory quinone and phosphatidylethanolamine, phosphatidylglycerol, two aminophospholipids, two phospholipids and four unidentified lipids as polar lipids. The DNA G+C content of the strain N9T was found to be 63 mol%. 16S rRNA gene sequence analysis indicated that Rhodobacter sphaeroides, Rhodobacter johrii, Pseudorhodobacter ferrugineus, Rhodobacter azotoformans, Rhodobacter ovatus and Pseudorhodobacter aquimaris were the nearest phylogenetic neighbours, with pair-wise sequence similarities of 95.43, 95.36, 94.24, 95.31, 95.60 and 94.74 % respectively. -

Distribution of Sulfate-Reducing Communities from Estuarine to Marine Bay Waters Yannick Colin, M
Distribution of Sulfate-Reducing Communities from Estuarine to Marine Bay Waters Yannick Colin, M. Goñi-Urriza, C. Gassie, E. Carlier, M. Monperrus, R. Guyoneaud To cite this version: Yannick Colin, M. Goñi-Urriza, C. Gassie, E. Carlier, M. Monperrus, et al.. Distribution of Sulfate- Reducing Communities from Estuarine to Marine Bay Waters. Microbial Ecology, Springer Verlag, 2017, 73 (1), pp.39-49. 10.1007/s00248-016-0842-5. hal-01499135 HAL Id: hal-01499135 https://hal.archives-ouvertes.fr/hal-01499135 Submitted on 26 Sep 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - ShareAlike| 4.0 International License Microb Ecol (2017) 73:39–49 DOI 10.1007/s00248-016-0842-5 MICROBIOLOGY OF AQUATIC SYSTEMS Distribution of Sulfate-Reducing Communities from Estuarine to Marine Bay Waters Yannick Colin 1,2 & Marisol Goñi-Urriza1 & Claire Gassie1 & Elisabeth Carlier 1 & Mathilde Monperrus3 & Rémy Guyoneaud1 Received: 23 May 2016 /Accepted: 17 August 2016 /Published online: 31 August 2016 # Springer Science+Business Media New York 2016 Abstract Estuaries are highly dynamic ecosystems in which gradient. The concentration of cultured sulfidogenic microor- freshwater and seawater mix together. -

Phylogenetic and Functional Characterization of Symbiotic Bacteria in Gutless Marine Worms (Annelida, Oligochaeta)
Phylogenetic and functional characterization of symbiotic bacteria in gutless marine worms (Annelida, Oligochaeta) Dissertation zur Erlangung des Grades eines Doktors der Naturwissenschaften -Dr. rer. nat.- dem Fachbereich Biologie/Chemie der Universität Bremen vorgelegt von Anna Blazejak Oktober 2005 Die vorliegende Arbeit wurde in der Zeit vom März 2002 bis Oktober 2005 am Max-Planck-Institut für Marine Mikrobiologie in Bremen angefertigt. 1. Gutachter: Prof. Dr. Rudolf Amann 2. Gutachter: Prof. Dr. Ulrich Fischer Tag des Promotionskolloquiums: 22. November 2005 Contents Summary ………………………………………………………………………………….… 1 Zusammenfassung ………………………………………………………………………… 2 Part I: Combined Presentation of Results A Introduction .…………………………………………………………………… 4 1 Definition and characteristics of symbiosis ...……………………………………. 4 2 Chemoautotrophic symbioses ..…………………………………………………… 6 2.1 Habitats of chemoautotrophic symbioses .………………………………… 8 2.2 Diversity of hosts harboring chemoautotrophic bacteria ………………… 10 2.2.1 Phylogenetic diversity of chemoautotrophic symbionts …………… 11 3 Symbiotic associations in gutless oligochaetes ………………………………… 13 3.1 Biogeography and phylogeny of the hosts …..……………………………. 13 3.2 The environment …..…………………………………………………………. 14 3.3 Structure of the symbiosis ………..…………………………………………. 16 3.4 Transmission of the symbionts ………..……………………………………. 18 3.5 Molecular characterization of the symbionts …..………………………….. 19 3.6 Function of the symbionts in gutless oligochaetes ..…..…………………. 20 4 Goals of this thesis …….…………………………………………………………..