A 1926 Supplement
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prokaryotic Community Successions and Interactions in Marine Biofilms
Prokaryotic community successions and interactions in marine biofilms: the key role of Flavobacteriia Thomas Pollet, Lyria Berdjeb, Cédric Garnier, Gaël Durrieu, Christophe Le Poupon, Benjamin Misson, Jean-François Briand To cite this version: Thomas Pollet, Lyria Berdjeb, Cédric Garnier, Gaël Durrieu, Christophe Le Poupon, et al.. Prokary- otic community successions and interactions in marine biofilms: the key role of Flavobacteriia. FEMS Microbiology Ecology, Wiley-Blackwell, 2018, 94 (6), 10.1093/femsec/fiy083. hal-02024255 HAL Id: hal-02024255 https://hal-amu.archives-ouvertes.fr/hal-02024255 Submitted on 2 Mar 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Prokaryotic community successions and interactions in marine biofilms: the key role of Flavobacteriia Thomas Pollet, Lyria Berdjeb, Cédric Garnier, Gaël Durrieu, Christophe Le Poupon, Benjamin Misson, Jean-François Briand To cite this version: Thomas Pollet, Lyria Berdjeb, Cédric Garnier, Gaël Durrieu, Christophe -

Bacterial Epibiotic Communities of Ubiquitous and Abundant Marine Diatoms Are Distinct in Short- and Long-Term Associations
fmicb-09-02879 December 1, 2018 Time: 14:0 # 1 ORIGINAL RESEARCH published: 04 December 2018 doi: 10.3389/fmicb.2018.02879 Bacterial Epibiotic Communities of Ubiquitous and Abundant Marine Diatoms Are Distinct in Short- and Long-Term Associations Klervi Crenn, Delphine Duffieux and Christian Jeanthon* CNRS, Sorbonne Université, Station Biologique de Roscoff, Adaptation et Diversité en Milieu Marin, Roscoff, France Interactions between phytoplankton and bacteria play a central role in mediating biogeochemical cycling and food web structure in the ocean. The cosmopolitan diatoms Thalassiosira and Chaetoceros often dominate phytoplankton communities in marine systems. Past studies of diatom-bacterial associations have employed community- level methods and culture-based or natural diatom populations. Although bacterial assemblages attached to individual diatoms represents tight associations little is known on their makeup or interactions. Here, we examined the epibiotic bacteria of 436 Thalassiosira and 329 Chaetoceros single cells isolated from natural samples and Edited by: collection cultures, regarded here as short- and long-term associations, respectively. Matthias Wietz, Epibiotic microbiota of single diatom hosts was analyzed by cultivation and by cloning- Alfred Wegener Institut, Germany sequencing of 16S rRNA genes obtained from whole-genome amplification products. Reviewed by: The prevalence of epibiotic bacteria was higher in cultures and dependent of the host Lydia Jeanne Baker, Cornell University, United States species. Culture approaches demonstrated that both diatoms carry distinct bacterial Bryndan Paige Durham, communities in short- and long-term associations. Bacterial epibonts, commonly University of Washington, United States associated with phytoplankton, were repeatedly isolated from cells of diatom collection *Correspondence: cultures but were not recovered from environmental cells. -

Article-Associated Bac- Teria and Colony Isolation in Soft Agar Medium for Bacteria Unable to Grow at the Air-Water Interface
Biogeosciences, 8, 1955–1970, 2011 www.biogeosciences.net/8/1955/2011/ Biogeosciences doi:10.5194/bg-8-1955-2011 © Author(s) 2011. CC Attribution 3.0 License. Diversity of cultivated and metabolically active aerobic anoxygenic phototrophic bacteria along an oligotrophic gradient in the Mediterranean Sea C. Jeanthon1,2, D. Boeuf1,2, O. Dahan1,2, F. Le Gall1,2, L. Garczarek1,2, E. M. Bendif1,2, and A.-C. Lehours3 1Observatoire Oceanologique´ de Roscoff, UMR7144, INSU-CNRS – Groupe Plancton Oceanique,´ 29680 Roscoff, France 2UPMC Univ Paris 06, UMR7144, Adaptation et Diversite´ en Milieu Marin, Station Biologique de Roscoff, 29680 Roscoff, France 3CNRS, UMR6023, Microorganismes: Genome´ et Environnement, Universite´ Blaise Pascal, 63177 Aubiere` Cedex, France Received: 21 April 2011 – Published in Biogeosciences Discuss.: 5 May 2011 Revised: 7 July 2011 – Accepted: 8 July 2011 – Published: 20 July 2011 Abstract. Aerobic anoxygenic phototrophic (AAP) bac- detected in the eastern basin, reflecting the highest diver- teria play significant roles in the bacterioplankton produc- sity of pufM transcripts observed in this ultra-oligotrophic tivity and biogeochemical cycles of the surface ocean. In region. To our knowledge, this is the first study to document this study, we applied both cultivation and mRNA-based extensively the diversity of AAP isolates and to unveil the ac- molecular methods to explore the diversity of AAP bacte- tive AAP community in an oligotrophic marine environment. ria along an oligotrophic gradient in the Mediterranean Sea By pointing out the discrepancies between culture-based and in early summer 2008. Colony-forming units obtained on molecular methods, this study highlights the existing gaps in three different agar media were screened for the production the understanding of the AAP bacteria ecology, especially in of bacteriochlorophyll-a (BChl-a), the light-harvesting pig- the Mediterranean Sea and likely globally. -

GHCMA Newsletter JUNE 2021 FINAL
In this edition: Projects continue within restrictions RCS consultation to begin and new funding welcomed Moyne River works begin Despite recent COVID-19 restrictions we have still been able More Bittern habitat protected to deliver some great outcomes with our partners in the Button Wrinkleworts survive first region, whilst some projects have been delayed or events summer in Dunkeld rescheduled. Spiny rice-flower planting day at This month’s newsletter has some stories of those Skipton partnerships, projects and achievements over the last DISA success in Hamilton month. There have been some great outcomes for our Vegetation monitoring field day threatened species such as the Australasian Bittern, Red- Estuary projects begin along the tailed Black Cockatoo, Button Wrinklewort and Spiny Riceflower, whilst the region continues to play a lead role coast with digital innovation in agriculture through hosting the Industry partnerships help second DISA Festival in Hamilton. cockatoos Victorian Landcare Awards open We were very pleased to hear that additional funding has been made available for RAMSAR, Flagship Waterways and for nominations Landcare in the recent State budget. This will be very welcome news for our partners, community groups and Glenelg Hopkins CMA Landcarers. www.ghcma.vic.gov.au From July 10 the public will have the opportunity to provide Telephone: (03) 5571 2526 feedback on the Regional Catchment Strategy. We welcome Email: [email protected] the feedback on the plan for land, water and biodiversity Postal: PO Box 502 management in our region through until 2027. Hamilton Victoria 3300 Adam Bester, CEO Glenelg Hopkins CMA CMA NEWS RCS public consultation period begins July 10 The period for public comment on the Glenelg Hopkins CMA Regional Catchment Strategy 2021- 2027 will begin on July 10, for four weeks. -

Reclassification of Agrobacterium Ferrugineum LMG 128 As Hoeflea
International Journal of Systematic and Evolutionary Microbiology (2005), 55, 1163–1166 DOI 10.1099/ijs.0.63291-0 Reclassification of Agrobacterium ferrugineum LMG 128 as Hoeflea marina gen. nov., sp. nov. Alvaro Peix,1 Rau´l Rivas,2 Martha E. Trujillo,2 Marc Vancanneyt,3 Encarna Vela´zquez2 and Anne Willems3 Correspondence 1Departamento de Produccio´n Vegetal, Instituto de Recursos Naturales y Agrobiologı´a, Encarna Vela´zquez IRNA-CSIC, Spain [email protected] 2Departamento de Microbiologı´a y Gene´tica, Lab. 209, Edificio Departamental, Campus Miguel de Unamuno, Universidad de Salamanca, 37007 Salamanca, Spain 3Laboratory of Microbiology, Dept Biochemistry, Physiology and Microbiology, Faculty of Sciences, Ghent University, Ghent, Belgium Members of the species Agrobacterium ferrugineum were isolated from marine environments. The type strain of this species (=LMG 22047T=ATCC 25652T) was recently reclassified in the new genus Pseudorhodobacter, in the order ‘Rhodobacterales’ of the class ‘Alphaproteobacteria’. Strain LMG 128 (=ATCC 25654) was also initially classified as belonging to the species Agrobacterium ferrugineum; however, the nearly complete 16S rRNA gene sequence of this strain indicated that it does not belong within the genus Agrobacterium or within the genus Pseudorhodobacter. The closest related organism, with 95?5 % 16S rRNA gene similarity, was Aquamicrobium defluvii from the family ‘Phyllobacteriaceae’ in the order ‘Rhizobiales’. The remaining genera from this order had 16S rRNA gene sequence similarities that were lower than 95?1 % with respect to strain LMG 128. These phylogenetic distances suggested that strain LMG 128 belonged to a different genus. The major fatty acid present in strain LMG 128 was mono-unsaturated straight chain 18 : 1v7c. -
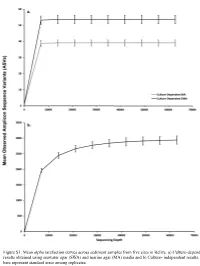
Figure S1. Mean Alpha Rarefaction Curves Across Sediment Samples from Five Sites in Belize
Figure S1. Mean alpha rarefaction curves across sediment samples from five sites in Belize. a) Culture-dependent results obtained using seawater agar (SWA) and marine agar (MA) media and b) Culture- independent results. Error bars represent standard error among replicates. a. 200 150 100 Faith’s PD Faith’s 50 0 Culturedependent MA Culturedependent SWA Cultureindependent Method b. 0.8 0.6 Pielou’s Evenness 0.4 Culturedependent MA Culturedependent SWA Cultureindependent Method Figure S2. Alpha diversity boxplots of marine sediment microbial communities from Carrie Bow Cay, Belize in culture-dependent and culture-independent samples determined using a) Faith’s Phylogenetic Diversity Index and b) Pielou’s Evenness. Culture-dependent methods include the use of marine agar medium (MA) and seawater agar medium (SWA. Data points are overlayed on the boxplot to show variation. a. 100% Proteobacteria Nitrospinae Bacteroidetes Fibrobacteres Planctomycetes Entotheonellaeota 90% Cyanobacteria Marinimicrobia (SAR406 clade) Firmicutes Deinococcus-Thermus Chloroflexi Margulisbacteria [A] Thaumarchaeota Unassigned 80% Acidobacteria Archaea UA Verrucomicrobia Acetothermia Actinobacteria Elusimicrobia 70% [A] Nanoarchaeaeota Tenericutes Kiritimatiellaeota TA06 Latescibacteria WS2 60% Spirochaetes Dependentiae Patescibacteria LCP-89 Gemmatimonadetes FCPU426 Omnitrophicaeota WPS-2 50% Fusobacteria CK-2C2-2 Bacteria UA Armatimonadetes [A] Euryarchaeota [A] Altiarchaeota Relative Percent 40% Calditrichaeota Poribacteria Lentisphaerae Cloacimonetes [A] Crenarchaeota -

Comparative Proteomic Profiling of Newly Acquired, Virulent And
www.nature.com/scientificreports OPEN Comparative proteomic profling of newly acquired, virulent and attenuated Neoparamoeba perurans proteins associated with amoebic gill disease Kerrie Ní Dhufaigh1*, Eugene Dillon2, Natasha Botwright3, Anita Talbot1, Ian O’Connor1, Eugene MacCarthy1 & Orla Slattery4 The causative agent of amoebic gill disease, Neoparamoeba perurans is reported to lose virulence during prolonged in vitro maintenance. In this study, the impact of prolonged culture on N. perurans virulence and its proteome was investigated. Two isolates, attenuated and virulent, had their virulence assessed in an experimental trial using Atlantic salmon smolts and their bacterial community composition was evaluated by 16S rRNA Illumina MiSeq sequencing. Soluble proteins were isolated from three isolates: a newly acquired, virulent and attenuated N. perurans culture. Proteins were analysed using two-dimensional electrophoresis coupled with liquid chromatography tandem mass spectrometry (LC–MS/MS). The challenge trial using naïve smolts confrmed a loss in virulence in the attenuated N. perurans culture. A greater diversity of bacterial communities was found in the microbiome of the virulent isolate in contrast to a reduction in microbial community richness in the attenuated microbiome. A collated proteome database of N. perurans, Amoebozoa and four bacterial genera resulted in 24 proteins diferentially expressed between the three cultures. The present LC–MS/ MS results indicate protein synthesis, oxidative stress and immunomodulation are upregulated in a newly acquired N. perurans culture and future studies may exploit these protein identifcations for therapeutic purposes in infected farmed fsh. Neoparamoeba perurans is an ectoparasitic protozoan responsible for the hyperplastic gill infection of marine cultured fnfsh referred to as amoebic gill disease (AGD)1. -

Diversity of Cultivated and Metabolically Active Aerobic Anoxygenic Phototrophic Bacteria Along an Oligotrophic Gradientthe in Mediterranean Sea C
Discussion Paper | Discussion Paper | Discussion Paper | Discussion Paper | Biogeosciences Discuss., 8, 4421–4457, 2011 Biogeosciences www.biogeosciences-discuss.net/8/4421/2011/ Discussions doi:10.5194/bgd-8-4421-2011 © Author(s) 2011. CC Attribution 3.0 License. This discussion paper is/has been under review for the journal Biogeosciences (BG). Please refer to the corresponding final paper in BG if available. Diversity of cultivated and metabolically active aerobic anoxygenic phototrophic bacteria along an oligotrophic gradient in the Mediterranean Sea C. Jeanthon1,2, D. Boeuf1,2, O. Dahan1,2, F. Le Gall1,2, L. Garczarek1,2, E. M. Bendif1,2, and A.-C. Lehours3 1INSU-CNRS, UMR 7144, Observatoire Oceanologique´ de Roscoff, Groupe Plancton Oceanique,´ 29680 Roscoff, France 2UPMC Univ Paris 06, UMR 7144, Adaptation et Diversite´ en Milieu Marin, Station Biologique de Roscoff, 29680 Roscoff, France 3CNRS, UMR 6023, Microorganismes: Genome´ et Environnement, Universite´ Blaise Pascal, 63177 Aubiere` Cedex, France Received: 21 April 2011 – Accepted: 29 April 2011 – Published: 5 May 2011 Correspondence to: C. Jeanthon (jeanthon@sb-roscoff.fr) Published by Copernicus Publications on behalf of the European Geosciences Union. 4421 Discussion Paper | Discussion Paper | Discussion Paper | Discussion Paper | Abstract Aerobic anoxygenic phototrophic (AAP) bacteria play significant roles in the bacterio- plankton productivity and biogeochemical cycles of the surface ocean. In this study, we applied both cultivation and mRNA-based molecular methods to explore the diversity of 5 AAP bacteria along an oligotrophic gradient in the Mediterranean Sea in early summer 2008. Colony-forming units obtained on three different agar media were screened for the production of bacteriochlorophyll-a (BChl-a), the light-harvesting pigment of AAP bacteria. -
Albirhodobacter Marinus Gen. Nov., Sp. Nov., a Member of the Family Rhodobacteriaceae Isolated from Sea Shore Water of Visakhapatnam, India
Author version: Antonie van Leeuwenhoek, vol.103; 2013; 347-355 Albirhodobacter marinus gen. nov., sp. nov., a member of the family Rhodobacteriaceae isolated from sea shore water of Visakhapatnam, India Nupur1, Bhumika, Vidya1., Srinivas, T. N. R2,3, Anil Kumar, P1* 1Microbial Type Culture Collection and Gene bank, Institute of Microbial Technology (CSIR), Sector 39A, Chandigarh - 160 036, INDIA 2National Institute of Oceanography (CSIR), Regional centre, P B No. 1913, Dr. Salim Ali Road, Kochi - 682018 (Kerala), INDIA Present Address: 3National Institute of Oceanography (CSIR), Regional centre, 176, Lawsons Bay Colony, Visakhapatnam - 530 017 (Andhra Pradesh), INDIA Address for correspondence* Dr. P. Anil Kumar Microbial Type Culture Collection and Gene bank Institute of Microbial Technology, Sector 39A, Chandigarh - 160 036, INDIA Email: [email protected] Phone: +91-172-6665170 1 Abstract A novel marine, Gram-negative, rod-shaped bacterium, designated strain N9T, was isolated from a water sample of the sea shore at Visakhapatnam, Andhra Pradesh (India). Strain N9T was found to be positive for oxidase and catalase activities. The fatty acids were found to be dominated by C16:0, C18:1 ω7c and summed in feature 3 (C16:1 ω7c and/or C16:1 ω6c). Strain N9T was determined to contain Q-10 as the major respiratory quinone and phosphatidylethanolamine, phosphatidylglycerol, two aminophospholipids, two phospholipids and four unidentified lipids as polar lipids. The DNA G+C content of the strain N9T was found to be 63 mol%. 16S rRNA gene sequence analysis indicated that Rhodobacter sphaeroides, Rhodobacter johrii, Pseudorhodobacter ferrugineus, Rhodobacter azotoformans, Rhodobacter ovatus and Pseudorhodobacter aquimaris were the nearest phylogenetic neighbours, with pair-wise sequence similarities of 95.43, 95.36, 94.24, 95.31, 95.60 and 94.74 % respectively. -

Discover the History of Warrnambool's Streets
Discover the history of Warrnambool's streets Street Name Description Locality Length Origin of Street Name Abbey Lane A laneway running between Hyland and Hart Streets, south of Timor Warrnambool 495m Benjamin Abbey (1862-1943) served two terms as Councillor 1913-16 and 1920-30. Served as Mayor 1924-26 during the Street. building of the Municipal Chambers. He was Manager of the Warrnambool branch of the Co-Operative Box Works of Victoria situated in South Warrnambool and a Trustee of the Methodist Church. His first wife Annie (nee Newman) died in Appears, unnamed, on an 1890 map. 1916 and his 2nd wife, Anastasia, died in 1994. This unnamed road was named Abbey Lane by the City of Warrnambool on 29th April 1991. The Council minutes and Government Gazette specifically name only the section between Hart and Hyland Streets which means the section between Hart and Ryot Streets is technically still unnamed. Aberline Road A northerly continuation of McKiernan Road, running from the Moore Warrnambool 1917m Joseph Aberline (1809-1874) arrived in Warrnambool in 1849 after spending some years in New Zealand. His property, Street/Dales Road intersection north to Wangoom Road. "The Grove", built on Wangoom Road in the 1860s was the site of a brick-making enterprise established by his son, John (1854-1940) in 1891. It was from the Wangoom Road property that large boulders were taken for use as some of the filling A very old road that appears on an 1856 map of Warrnambool. for the Warrnambool breakwater. Some older maps call it Aberlines Road. -

Roseovarius Azorensis Sp. Nov., Isolated from Seawater At
Author version: Antonie van Leeuwenhoek, vol.105(3); 2014; 571-578 Roseovarius azorensis sp. nov., isolated from seawater at Espalamaca, Azores Raju Rajasabapathy • Chellandi Mohandass • Syed Gulam Dastager • Qing Liu • Thi-Nhan Khieu • Chu Ky Son • Wen-Jun Li • Ana Colaco Raju Rajasabapathy · Chellandi Mohandass* Biological Oceanography Division, CSIR-National Institute of Oceanography, Dona Paula, Goa 403 004, India. E-mail: [email protected] Syed Gulam Dastager NCIM Resource Center, CSIR-National Chemical Laboratory, Dr. Homi Bhabha road, Pune 411 008, India Qing Liu · Thi-Nhan Khieu · Wen-Jun Li Yunnan Institute of Microbiology, Yunnan University, Kunming, Yunnan 650091, P.R. China Thi-Nhan Khieu · Chu Ky Son School of Biotechnology and Food Technology, Hanoi University of Science and Technology, Vietnam Ana Colaco IMAR-Department of Oceanography and Fisheries, University Açores, Cais de Sta Cruz, 9901-862, Horta, Portugal Abstract A Gram-negative, motile, non-spore forming, rod shaped aerobic bacterium, designated strain SSW084T, was isolated from a surface seawater sample collected at Espalamaca (38°33’N; 28°39’W), Azores. Growth was found to occur from 15 – 40 °C (optimum 30 °C), at pH 7.0 – 9.0 (optimum pH 7.0) and with 25 to 100 % seawater or 0.5 – 7.0 % NaCl in the presence of Mg2+ and Ca2+; no growth was found with NaCl alone. Colonies on seawater nutrient agar (SWNA) were observed to be punctiform, white, convex, circular, smooth, and translucent. Strain SSW084T did not grow on Zobell Marine Agar (ZMA) and tryptic soy agar (TSA) even when seawater supplemented. The major respiratory quinone was found to be Q-10 and the G+C content was determined to be 61.9 mol%. -

Moyne Shire Emergency Managament Plan
Moyne Shire FLOOD EMERGENCY PLAN A Sub-Plan of the Municipal Emergency Management Plan For Moyne Shire Council and VICSES Port Fairy, Mortlake, Warrnambool, Terang and Port Campbell Units Version 3, March 2021 Table of Contents Part 1. Introduction ....................................................................................................................................... 1 1.1 Approval and Endorsement .................................................................................................................... 1 1.2 Purpose and Scope of this Flood Emergency Plan ................................................................................ 2 1.3 Responsibility for Planning, Review & Maintenance of this Plan ........................................................... 2 Part 2. BEFORE: Prevention / preparedness arrangements .................................................................... 3 2.1 Community Engagement and Awareness .............................................................................................. 3 2.2 Structural Flood Mitigation Measures ..................................................................................................... 3 2.3 Non-structural Flood Mitigation Measures .............................................................................................. 3 2.3.1 Exercising the Plan ................................................................................................................................. 3 2.3.2 Flood Warning .......................................................................................................................................