Hippotragus Leucophaeus)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hippotragus Equinus – Roan Antelope
Hippotragus equinus – Roan Antelope authorities as there may be no significant genetic differences between the two. Many of the Roan Antelope in South Africa are H. e. cottoni or equinus x cottoni (especially on private properties). Assessment Rationale This charismatic antelope exists at low density within the assessment region, occurring in savannah woodlands and grasslands. Currently (2013–2014), there are an observed 333 individuals (210–233 mature) existing on nine formally protected areas within the natural distribution range. Adding privately protected subpopulations and an Cliff & Suretha Dorse estimated 0.8–5% of individuals on wildlife ranches that may be considered wild and free-roaming, yields a total mature population of 218–294 individuals. Most private Regional Red List status (2016) Endangered subpopulations are intensively bred and/or kept in camps C2a(i)+D*†‡ to exclude predators and to facilitate healthcare. Field National Red List status (2004) Vulnerable D1 surveys are required to identify potentially eligible subpopulations that can be included in this assessment. Reasons for change Non-genuine: While there was an historical crash in Kruger National Park New information (KNP) of 90% between 1986 and 1993, the subpopulation Global Red List status (2008) Least Concern has since stabilised at c. 50 individuals. Overall, over the past three generations (1990–2015), based on available TOPS listing (NEMBA) Vulnerable data for nine formally protected areas, there has been a CITES listing None net population reduction of c. 23%, which indicates an ongoing decline but not as severe as the historical Endemic Edge of Range reduction. Further long-term data are needed to more *Watch-list Data †Watch-list Threat ‡Conservation Dependent accurately estimate the national population trend. -

Understanding the Behavioural Trade-Offs Made by Blue Wildebeest (Connochaetes Taurinus): the Importance of Resources, Predation and the Landscape
Understanding the behavioural trade-offs made by blue wildebeest (Connochaetes taurinus): the importance of resources, predation and the landscape Rebecca Dannock Bachelor of Science (Hons) in Zoology A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2016 School of Biological Sciences Abstract Prey individuals must constantly make decisions regarding safety and resource acquisition to ensure that they acquire enough resources without being predated upon. These decisions result in a trade-off between resource acquisition behaviours (such as foraging and drinking) and safety behaviours (such as grouping and vigilance). This trade- off is likely to be affected by the social and environmental factors that an individual experiences, including the individual’s location in the landscape. The overall objective of my PhD was to understand the decisions a migratory ungulate makes in order to acquire enough resources, while not becoming prey, and to understand how these decisions are influenced by social and environmental factors. In order to do this, I studied the behaviour of blue wildebeest (Connochaetes taurinus) in Etosha National Park, Namibia, between 2013 and 2015. I studied wildebeests’ behaviour while they acquired food and water and moved within the landscape. Along with observational studies, I also used lion (Panthera leo) roar playbacks to experimentally manipulate perceived predator presence to test wildebeests’ responses to immediate predation risk. For Chapter 2 I studied the foraging-vigilance trade-off of wildebeest to determine how social and environmental factors, including the location within the landscape, were correlated with wildebeests’ time spent foraging and vigilant as well as their bite rate. -

Sustainable Wildlife Management
249ISSN 0041-6436 An international journal of forestry and forest industries Vol. 68 2017/1 SUSTAINABLE WILDLIFE MANAGEMENT 249ISSN 0041-6436 An international journal of forestry and forest industries Vol. 68 2017/1 Editor: A. Sarre Editorial Advisory Board: S. Braatz, I. Buttoud, P. Csoka, L. Flejzor, T. Hofer, Contents F. Kafeero, W. Kollert, S. Lapstun, D. Mollicone, D. Reeb, S. Rose, J. Tissari, Editorial 2 P. van Lierop Emeritus Advisers: J. Ball, I.J. Bourke, R. Cooney, C. Freese, H. Dublin, D. Roe, D. Mallon, M. Knight, C. Palmberg-Lerche, L. Russo R. Emslie, M. Pani, V. Booth, S. Mahoney and C. Buyanaa Regional Advisers: F. Bojang, P. Durst, The baby and the bathwater: trophy hunting, conservation A.A. Hamid, J. Meza and rural livelihoods 3 Unasylva is published in English, French J. Stahl and T. De Meulenaer and Spanish. Subscriptions can be obtained CITES and the international trade in wildlife 17 by sending an e-mail to [email protected]. Subscription requests from institutions Y. Vizina and D. Kobei (e.g. libraries, companies, organizations, Indigenous peoples and sustainable wildlife management universities) rather than individuals are in the global era 27 preferred in order to make the journal accessible to more readers. D. Roe, R. Cooney, H. Dublin, D. Challender, D. Biggs, D. Skinner, All issues of Unasylva are available online M. Abensperg-Traun, N. Ahlers, R. Melisch and M. Murphree free of charge at www.fao.org/forestry/ First line of defence: engaging communities in tackling unasylva. Comments and queries are welcome: wildlife crime 33 [email protected]. -

Mixed-Species Exhibits with Pigs (Suidae)
Mixed-species exhibits with Pigs (Suidae) Written by KRISZTIÁN SVÁBIK Team Leader, Toni’s Zoo, Rothenburg, Luzern, Switzerland Email: [email protected] 9th May 2021 Cover photo © Krisztián Svábik Mixed-species exhibits with Pigs (Suidae) 1 CONTENTS INTRODUCTION ........................................................................................................... 3 Use of space and enclosure furnishings ................................................................... 3 Feeding ..................................................................................................................... 3 Breeding ................................................................................................................... 4 Choice of species and individuals ............................................................................ 4 List of mixed-species exhibits involving Suids ........................................................ 5 LIST OF SPECIES COMBINATIONS – SUIDAE .......................................................... 6 Sulawesi Babirusa, Babyrousa celebensis ...............................................................7 Common Warthog, Phacochoerus africanus ......................................................... 8 Giant Forest Hog, Hylochoerus meinertzhageni ..................................................10 Bushpig, Potamochoerus larvatus ........................................................................ 11 Red River Hog, Potamochoerus porcus ............................................................... -

List of 28 Orders, 129 Families, 598 Genera and 1121 Species in Mammal Images Library 31 December 2013
What the American Society of Mammalogists has in the images library LIST OF 28 ORDERS, 129 FAMILIES, 598 GENERA AND 1121 SPECIES IN MAMMAL IMAGES LIBRARY 31 DECEMBER 2013 AFROSORICIDA (5 genera, 5 species) – golden moles and tenrecs CHRYSOCHLORIDAE - golden moles Chrysospalax villosus - Rough-haired Golden Mole TENRECIDAE - tenrecs 1. Echinops telfairi - Lesser Hedgehog Tenrec 2. Hemicentetes semispinosus – Lowland Streaked Tenrec 3. Microgale dobsoni - Dobson’s Shrew Tenrec 4. Tenrec ecaudatus – Tailless Tenrec ARTIODACTYLA (83 genera, 142 species) – paraxonic (mostly even-toed) ungulates ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BOVIDAE (46 genera) - cattle, sheep, goats, and antelopes 1. Addax nasomaculatus - Addax 2. Aepyceros melampus - Impala 3. Alcelaphus buselaphus - Hartebeest 4. Alcelaphus caama – Red Hartebeest 5. Ammotragus lervia - Barbary Sheep 6. Antidorcas marsupialis - Springbok 7. Antilope cervicapra – Blackbuck 8. Beatragus hunter – Hunter’s Hartebeest 9. Bison bison - American Bison 10. Bison bonasus - European Bison 11. Bos frontalis - Gaur 12. Bos javanicus - Banteng 13. Bos taurus -Auroch 14. Boselaphus tragocamelus - Nilgai 15. Bubalus bubalis - Water Buffalo 16. Bubalus depressicornis - Anoa 17. Bubalus quarlesi - Mountain Anoa 18. Budorcas taxicolor - Takin 19. Capra caucasica - Tur 20. Capra falconeri - Markhor 21. Capra hircus - Goat 22. Capra nubiana – Nubian Ibex 23. Capra pyrenaica – Spanish Ibex 24. Capricornis crispus – Japanese Serow 25. Cephalophus jentinki - Jentink's Duiker 26. Cephalophus natalensis – Red Duiker 1 What the American Society of Mammalogists has in the images library 27. Cephalophus niger – Black Duiker 28. Cephalophus rufilatus – Red-flanked Duiker 29. Cephalophus silvicultor - Yellow-backed Duiker 30. Cephalophus zebra - Zebra Duiker 31. Connochaetes gnou - Black Wildebeest 32. Connochaetes taurinus - Blue Wildebeest 33. Damaliscus korrigum – Topi 34. -
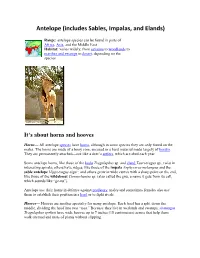
Antelope (Includes Sables, Impalas, and Elands)
Antelope (includes Sables, Impalas, and Elands) Range: antelope species can be found in parts of Africa, Asia, and the Middle East Habitat: varies widely, from savanna to woodlands to marshes and swamps to desert, depending on the species It’s about horns and hooves Horns— All antelope species have horns, although in some species they are only found on the males. The horns are made of a bony core, encased in a hard material made largely of keratin. They are permanently attached—not like a deer’s antlers, which are shed each year. Some antelope horns, like those of the kudu Tragelaphus sp. and eland Taurotragus sp., twist in interesting spirals; others have ridges, like those of the impala Aephyceros melampus and the sable antelope Hippotragus niger; and others grow in wide curves with a sharp point on the end, like those of the wildebeest Connochaetes sp. (also called the gnu, a name it gets from its call, which sounds like “ge-nu”). Antelope use their horns in defense against predators; males and sometimes females also use them to establish their position in a herd or to fight rivals Hooves— Hooves are another specialty for many antelope. Each hoof has a split down the middle, dividing the hoof into two “toes.” Because they live in wetlands and swamps, sitatungas Tragelaphus spekeii have wide hooves up to 7 inches (18 centimeters) across that help them walk on mud and mats of plants without slipping. Nile lechwes Kobus magaceros, which also live in swampy areas, have long, pointed hooves to give them sure footing in the water. -

The Practical Side of Immunocontraception: Zona Proteins and Wildlife
This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues. Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited. In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier’s archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright Author's personal copy Journal of Reproductive Immunology 83 (2009) 151–157 Contents lists available at ScienceDirect Journal of Reproductive Immunology journal homepage: www.elsevier.com/locate/jreprimm The practical side of immunocontraception: zona proteins and wildlife J.F. Kirkpatrick a,∗, A. Rowan b, N. Lamberski c, R. Wallace d, K. Frank a, R. Lyda a a The Science and Conservation Center, ZooMontana, Billings, MT 59106, USA b The Humane Society of the United States, Gaithersburg, MD, USA c San Diego Wild Animal Park, Escondido, CA, USA d Milwaukee County Zoo, Milwaukee, WI, USA article info abstract Article history: With shrinking habitat, the humane control of certain wildlife populations is relevant. The Received 31 December 2008 contraceptive vaccine based on native porcine zona pellucida (PZP) has been applied to Received in revised form 16 June 2009 various wildlife populations for 20 years. Prominent efforts include wild horses, urban Accepted 16 June 2009 deer, zoo animals and African elephants, among others. -

Antelopes, Gazelles, Cattle, Goats, Sheep, and Relatives
© Copyright, Princeton University Press. No part of this book may be distributed, posted, or reproduced in any form by digital or mechanical means without prior written permission of the publisher. INTRODUCTION RECOGNITION The family Bovidae, which includes Antelopes, Cattle, Duikers, Gazelles, Goats, and Sheep, is the largest family within Artiodactyla and the most diverse family of ungulates, with more than 270 recent species. Their common characteristic is their unbranched, non-deciduous horns. Bovids are primarily Old World in their distribution, although a few species are found in North America. The name antelope is often used to describe many members of this family, but it is not a definable, taxonomically based term. Shape, size, and color: Bovids encompass an extremely wide size range, from the minuscule Royal Antelope and the Dik-diks, weighing as little as 2 kg and standing 25 to 35 cm at the shoulder, to the Asian Wild Water Buffalo, which weighs as much as 1,200 kg, and the Gaur, which measures up to 220 cm at the shoulder. Body shape varies from relatively small, slender-limbed, and thin-necked species such as the Gazelles to the massive, stocky wild cattle (fig. 1). The forequarters may be larger than the hind, or the reverse, as in smaller species inhabiting dense tropical forests (e.g., Duikers). There is also a great variety in body coloration, although most species are some shade of brown. It can consist of a solid shade, or a patterned pelage. Antelopes that rely on concealment to avoid predators are cryptically colored. The stripes and blotches seen on the hides of Bushbuck, Bongo, and Kudu also function as camouflage by helping to disrupt the animals’ outline. -

Species and Genotypes of Cryptosporidium RON FAYER USDA AGRICULTURAL RESEARCH SERVICE WHAT IS a SPECIES?
Species and Genotypes of Cryptosporidium RON FAYER USDA AGRICULTURAL RESEARCH SERVICE WHAT IS A SPECIES? It is a basic unit of biological classification and a taxonomic rank. A species is often defined as a group of organisms capable of interbreeding and producing fertile offspring. More precise or differing measures can be used, such as similarity of DNA, morphology or ecological niche. Species that are believed to have the same ancestors are grouped together, as a genus. All species have two part name (a "binomial name"). The first part of a binomial name is the genus of the species. The second part is the specific name. For example, Boa constrictor which is commonly called by its binomial name, and is one of four species of the Boa genus. The first part of the name is capitalized, and the second part has a lower case. The two part name is written in italics. What are the sources of Cryptosporidium? FIELD MOUSE WOODCHUCK VOLE SKUNK BEAVER More than 150 species of animals infected with Cryptosporidium spp. Order Artiodactyla Addax nasomaculatus (Addax) Aepyceros melampus (Impala) Cervus elaphus (Red deer/elk/wapiti) Ammotragus lervia (Barbary sheep) Cervus eldi (Eld's deer) Alces alces (moose) Cervus nippon (Sika deer) Antidorcas marsupialis (Springbok) Cervus unicolor (Sambar) Antilocapra americana (Pronghorn) Connochaetes gnou (Wildebeest) Antilope cervicapra (Blackbuck) Connochaetes taurinus (Blue-eared gnu) Axis axis (Axis deer) Dama dama (Fallow deer) Bison bison (American bison) Elaphurus davidianus (Pere David’s deer) Bison bonasus -

Sable Antelope (Hippotragus Niger)
Sable Antelope (Hippotragus Niger) Kingdom: Animalia Phylum: Chordata Class: Mammalia Order: Artiodactyla Family: Bovidae WHAT IT IS The name "sable" refers to the black color of the male's coat. Males are about 9 feet long with a shoulder height of 4 feet. The male weighs about 500 pounds and is 20 percent larger than the female. The male's coat is dark brown to black with white under parts, while the female and young males have rich russet coats. Both males and females have stout, scythe-like ringed horns that grow to a length of 20-60 inches and both have stiff manes. Sable antelopes have large ears, large eyes and a keen sense of hearing, vision and smell. WHERE IT LIVES Southern Savanna, from southeastern Kenya, eastern Tanzania, and Mozambique to Angola and southern Zaire, mainly in the Miombo Woodland Zone. The giant sable, isolated and vulnerable in central Angola, is one of the most endangered antelopes. ECOLOGY Preferred habitat combines savanna woodland and grassland; trees (fire- resistant, broad-leafed, and deciduous) widely spaced with understory of sparse grasses utilized in rainy season. Drainage-line and floodplain grasslands that produce new growth after the annual fires keep sable in open during dry season. A grazer/browser eats grasses supplemented by foliage and herbs, especially kinds growing on woodland termite mounds. Goes to water at least every other day and regularly visits salt licks (also chews bones to make up for mineral-deficient soils). ACTIVITY Traveling on average approximately 1/2mi a day, sables are especially sedentary on dry-season pastures, when herds may spend weeks on the same field, leaving only to go to water or seek shade during hottest hours. -

The Introduction of Alien Mammals Into the Broader Western and Northern Cape Provinces of South Africa
The introduction of alien mammals into the broader Western and Northern Cape Provinces of South Africa A facsimile extract from Skead (2011) Graham Kerley and André Boshoff Centre for African Conservation Ecology Report No. 62 Nelson Mandela Metropolitan University April 2015 PO Box 77000 Port Elizabeth South Africa Introduction The ongoing debate on the occurrence, desirability and consequences of the introduction of alien (i.e. those species that do not occur in an area naturally) mammals into areas of South Africa (Castley et al. 2001, Cousins et al. 2010) highlights the need for a robust historical perspective on such introductions. Thus we need to know which species were introduced, when and where this happened, and also why such introductions took place. Alas, for most of South Africa there is a paucity of such information, as both the authorities responsible for the relevant policy and its application, as well as those who undertook these introductions, failed to maintain an accurate record; and the records that do exist are typically buried in obscure archives. As a consequence, we are left with a patchy, frequently romanticised, record of introductions of mammals that is of little help in explaining where and how (e.g. whether a species is free ranging or maintained in farmed system) alien mammals are currently distributed in South Africa. There is, however, one attempt to compile such an historical perspective, this for the broader Western Cape and Northern Cape provinces of South Africa. This material is published as a chapter in a book (Skead 2011) that may not be generally accessible. -

A Meta-Analysis of Human–Wildlife Conflict: South African and Global Perspectives
Sustainability 2017, 9, 34; doi:10.3390/su9010034 S1 of S11 Supplementary Materials: A Meta-Analysis of Human–Wildlife Conflict: South African and Global Perspectives Nimmi Seoraj-Pillai and Neville Pillay Table S1. Description of categories that gauge vulnerability of human-wildlife-conflict species and severity of conflict. A description of how species were categorised for vulnerability and conflict status is provided using guidelines proposed by Gittleman et al. (2001) and Inskip and Zimmermann (2009). These categories identified levels of biodiversity extinction vulnerability with corresponding abbreviations for such classification. Category Description Category Description Index of Vulnerability Conflict Status Animal appears only Wild animal rarely attacks people, Poorly researched, Low-scale once in the published seldom depredates livestock or crops, data deficient (PR) conflict (LSC) literature database rarely experiences retaliatory killing Animal appears two to Wild animal rarely attacks people, or Moderately four times in the Moderate-scale may frequently depredate livestock or persecuted (MP) database and may be conflict (MSC) crops, or experiences frequent moderately persecuted retaliatory killings Animal appears more Wild animal frequently attacks Severely than four times in the High-scale people and/or recurrently depredates persecuted (SP) database and may be conflict (HSC) livestock or crops, experiences severely persecuted frequent retaliatory killings Research required No research has been Anecdotal evidence of conflict is Status unknown (RR) or Future conducted on this species available. No scientific evidence in (SU) research (FR) in published literature literature Table S2. Problem animals that affected commercial farmers, local communities, subsistence farmers and pooled-farmers (subsistence and commercial farmers). Numbers denote the number of cases that appeared in the published literature database.