X Chromosome Evolution in Cetartiodactyla
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Camelids: New Players in the International Animal Production Context
Tropical Animal Health and Production (2020) 52:903–913 https://doi.org/10.1007/s11250-019-02197-2 REVIEWS Camelids: new players in the international animal production context Mousa Zarrin1 & José L. Riveros2 & Amir Ahmadpour1,3 & André M. de Almeida4 & Gaukhar Konuspayeva5 & Einar Vargas- Bello-Pérez6 & Bernard Faye7 & Lorenzo E. Hernández-Castellano8 Received: 30 October 2019 /Accepted: 22 December 2019 /Published online: 2 January 2020 # Springer Nature B.V. 2020 Abstract The Camelidae family comprises the Bactrian camel (Camelus bactrianus), the dromedary camel (Camelus dromedarius), and four species of South American camelids: llama (Lama glama),alpaca(Lama pacos)guanaco(Lama guanicoe), and vicuña (Vicugna vicugna). The main characteristic of these species is their ability to cope with either hard climatic conditions like those found in arid regions (Bactrian and dromedary camels) or high-altitude landscapes like those found in South America (South American camelids). Because of such interesting physiological and adaptive traits, the interest for these animals as livestock species has increased considerably over the last years. In general, the main animal products obtained from these animals are meat, milk, and hair fiber, although they are also used for races and work among other activities. In the near future, climate change will likely decrease agricultural areas for animal production worldwide, particularly in the tropics and subtropics where competition with crops for human consumption is a major problem already. In such conditions, extensive animal production could be limited in some extent to semi-arid rangelands, subjected to periodical draughts and erratic patterns of rainfall, severely affecting conventional livestock production, namely cattle and sheep. -

A Giant's Comeback
W INT E R 2 0 1 0 n o s d o D y l l i B Home to elephants, rhinos and more, African Heartlands are conservation landscapes large enough to sustain a diversity of species for centuries to come. In these landscapes— places like Kilimanjaro and Samburu—AWF and its partners are pioneering lasting conservation strate- gies that benefit wildlife and people alike. Inside TH I S ISSUE n e s r u a L a n a h S page 4 These giraffes are members of the only viable population of West African giraffe remaining in the wild. A few herds live in a small AWF Goes to West Africa area in Niger outside Regional Parc W. AWF launches the Regional Parc W Heartland. A Giant’s Comeback t looks like a giraffe, walks like a giraffe, eats totaling a scant 190-200 individuals. All live in like a giraffe and is indeed a giraffe. But a small area—dubbed “the Giraffe Zone”— IGiraffa camelopardalis peralta (the scientific outside the W National Parc in Niger, one of name for the West African giraffe) is a distinct the three national parks that lie in AWF’s new page 6 subspecies of mother nature’s tallest mammal, transboundary Heartland in West Africa (see A Quality Brew having split from a common ancestral popula- pp. 4-5). Conserving the slopes of Mt. Kilimanjaro tion some 35,000 years ago. This genetic Entering the Zone with good coffee. distinction is apparent in its large orange- Located southeast of Niamey, Niger’s brown skin pattern, which is more lightly- capital, the Giraffe Zone spans just a few hun- colored than that of other giraffes. -

Fitzhenry Yields 2016.Pdf
Stellenbosch University https://scholar.sun.ac.za ii DECLARATION By submitting this dissertation electronically, I declare that the entirety of the work contained therein is my own, original work, that I am the sole author thereof (save to the extent explicitly otherwise stated), that reproduction and publication thereof by Stellenbosch University will not infringe any third party rights and that I have not previously in its entirety or in part submitted it for obtaining any qualification. Date: March 2016 Copyright © 2016 Stellenbosch University All rights reserved Stellenbosch University https://scholar.sun.ac.za iii GENERAL ABSTRACT Fallow deer (Dama dama), although not native to South Africa, are abundant in the country and could contribute to domestic food security and economic stability. Nonetheless, this wild ungulate remains overlooked as a protein source and no information exists on their production potential and meat quality in South Africa. The aim of this study was thus to determine the carcass characteristics, meat- and offal-yields, and the physical- and chemical-meat quality attributes of wild fallow deer harvested in South Africa. Gender was considered as a main effect when determining carcass characteristics and yields, while both gender and muscle were considered as main effects in the determination of physical and chemical meat quality attributes. Live weights, warm carcass weights and cold carcass weights were higher (p < 0.05) in male fallow deer (47.4 kg, 29.6 kg, 29.2 kg, respectively) compared with females (41.9 kg, 25.2 kg, 24.7 kg, respectively), as well as in pregnant females (47.5 kg, 28.7 kg, 28.2 kg, respectively) compared with non- pregnant females (32.5 kg, 19.7 kg, 19.3 kg, respectively). -

Cuticle and Cortical Cell Morphology of Alpaca and Other Rare Animal Fibres
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Repositorio Institucional Universidad Nacional Autónoma de Chota The Journal of The Textile Institute ISSN: 0040-5000 (Print) 1754-2340 (Online) Journal homepage: http://www.tandfonline.com/loi/tjti20 Cuticle and cortical cell morphology of alpaca and other rare animal fibres B. A. McGregor & E. C. Quispe Peña To cite this article: B. A. McGregor & E. C. Quispe Peña (2017): Cuticle and cortical cell morphology of alpaca and other rare animal fibres, The Journal of The Textile Institute, DOI: 10.1080/00405000.2017.1368112 To link to this article: http://dx.doi.org/10.1080/00405000.2017.1368112 Published online: 18 Sep 2017. Submit your article to this journal Article views: 7 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=tjti20 Download by: [181.64.24.124] Date: 25 September 2017, At: 13:39 THE JOURNAL OF THE TEXTILE INSTITUTE, 2017 https://doi.org/10.1080/00405000.2017.1368112 Cuticle and cortical cell morphology of alpaca and other rare animal fibres B. A. McGregora and E. C. Quispe Peñab aInstitute for Frontier Materials, Deakin University, Geelong, Australia; bNational University Autonoma de Chota, Chota, Peru ABSTRACT ARTICLE HISTORY The null hypothesis of the experiments reported is that the cuticle and cortical morphology of rare Received 6 March 2017 animal fibres are similar. The investigation also examined if the productivity and age of alpacas were Accepted 11 August 2017 associated with cuticle morphology and if seasonal nutritional conditions were related to cuticle scale KEYWORDS frequency. -

Last Interglacial (MIS 5) Ungulate Assemblage from the Central Iberian Peninsula: the Camino Cave (Pinilla Del Valle, Madrid, Spain)
Palaeogeography, Palaeoclimatology, Palaeoecology 374 (2013) 327–337 Contents lists available at SciVerse ScienceDirect Palaeogeography, Palaeoclimatology, Palaeoecology journal homepage: www.elsevier.com/locate/palaeo Last Interglacial (MIS 5) ungulate assemblage from the Central Iberian Peninsula: The Camino Cave (Pinilla del Valle, Madrid, Spain) Diego J. Álvarez-Lao a,⁎, Juan L. Arsuaga b,c, Enrique Baquedano d, Alfredo Pérez-González e a Área de Paleontología, Departamento de Geología, Universidad de Oviedo, C/Jesús Arias de Velasco, s/n, 33005 Oviedo, Spain b Centro Mixto UCM-ISCIII de Evolución y Comportamiento Humanos, C/Sinesio Delgado, 4, 28029 Madrid, Spain c Departamento de Paleontología, Facultad de Ciencias Geológicas, Universidad Complutense de Madrid, Ciudad Universitaria, 28040 Madrid, Spain d Museo Arqueológico Regional de la Comunidad de Madrid, Plaza de las Bernardas, s/n, 28801-Alcalá de Henares, Madrid, Spain e Centro Nacional de Investigación sobre la Evolución Humana (CENIEH), Paseo Sierra de Atapuerca, s/n, 09002 Burgos, Spain article info abstract Article history: The fossil assemblage from the Camino Cave, corresponding to the late MIS 5, constitutes a key record to un- Received 2 November 2012 derstand the faunal composition of Central Iberia during the last Interglacial. Moreover, the largest Iberian Received in revised form 21 January 2013 fallow deer fossil population was recovered here. Other ungulate species present at this assemblage include Accepted 31 January 2013 red deer, roe deer, aurochs, chamois, wild boar, horse and steppe rhinoceros; carnivores and Neanderthals Available online 13 February 2013 are also present. The origin of the accumulation has been interpreted as a hyena den. Abundant fallow deer skeletal elements allowed to statistically compare the Camino Cave fossils with other Keywords: Early Late Pleistocene Pleistocene and Holocene European populations. -

The Status and Distribution of Mediterranean Mammals
THE STATUS AND DISTRIBUTION OF MEDITERRANEAN MAMMALS Compiled by Helen J. Temple and Annabelle Cuttelod AN E AN R R E IT MED The IUCN Red List of Threatened Species™ – Regional Assessment THE STATUS AND DISTRIBUTION OF MEDITERRANEAN MAMMALS Compiled by Helen J. Temple and Annabelle Cuttelod The IUCN Red List of Threatened Species™ – Regional Assessment The designation of geographical entities in this book, and the presentation of material, do not imply the expression of any opinion whatsoever on the part of IUCN or other participating organizations, concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. The views expressed in this publication do not necessarily reflect those of IUCN or other participating organizations. Published by: IUCN, Gland, Switzerland and Cambridge, UK Copyright: © 2009 International Union for Conservation of Nature and Natural Resources Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holder provided the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited without prior written permission of the copyright holder. Red List logo: © 2008 Citation: Temple, H.J. and Cuttelod, A. (Compilers). 2009. The Status and Distribution of Mediterranean Mammals. Gland, Switzerland and Cambridge, UK : IUCN. vii+32pp. ISBN: 978-2-8317-1163-8 Cover design: Cambridge Publishers Cover photo: Iberian lynx Lynx pardinus © Antonio Rivas/P. Ex-situ Lince Ibérico All photographs used in this publication remain the property of the original copyright holder (see individual captions for details). -

A Genus-Level Phylogenetic Analysis of Antilocapridae And
A GENUS-LEVEL PHYLOGENETIC ANALYSIS OF ANTILOCAPRIDAE AND IMPLICATIONS FOR THE EVOLUTION OF HEADGEAR MORPHOLOGY AND PALEOECOLOGY by HOLLEY MAY FLORA A THESIS Presented to the Department of EArth Sciences And the Graduate School of the University of Oregon in partiAl fulfillment of the requirements for the degree of MAster of Science September 2019 THESIS APPROVAL PAGE Student: Holley MAy Flora Title: A Genus-level Phylogenetic Analysis of AntilocApridae and ImplicAtions for the Evolution of HeAdgeAr Morphology and PAleoecology This Thesis has been accepted and approved in partiAl fulfillment of the requirements for the MAster of Science degree in the Department of EArth Sciences by: EdwArd Byrd DAvis Advisor SAmAntha S. B. Hopkins Core Member Matthew Polizzotto Core Member Stephen Frost Institutional RepresentAtive And JAnet Woodruff-Borden DeAn of the Graduate School Original Approval signatures are on file with the University of Oregon Graduate School Degree awArded September 2019. ii ã 2019 Holley MAy Flora This work is licensed under a CreAtive Commons Attribution-NonCommercial-NoDerivs (United States) License. iii THESIS ABSTRACT Holley MAy Flora MAster of Science Department of EArth Sciences September 2019 Title: A Genus-level Phylogenetic Analysis of AntilocApridae and ImplicAtions for the Evolution of HeAdgeAr Morphology and PAleoecology The shapes of Artiodactyl heAdgeAr plAy key roles in interactions with their environment and eAch other. Consequently, heAdgeAr morphology cAn be used to predict behavior. For eXAmple, lArger, recurved horns are typicAl of gregarious, lArge-bodied AnimAls fighting for mAtes. SmAller spike-like horns are more characteristic of small- bodied, paired mAtes from closed environments. Here, I report a genus-level clAdistic Analysis of the extinct family, AntilocApridae, testing prior hypotheses of evolutionary history And heAdgeAr evolution. -

Red List of Bangladesh Volume 2: Mammals
Red List of Bangladesh Volume 2: Mammals Lead Assessor Mohammed Mostafa Feeroz Technical Reviewer Md. Kamrul Hasan Chief Technical Reviewer Mohammad Ali Reza Khan Technical Assistants Selina Sultana Md. Ahsanul Islam Farzana Islam Tanvir Ahmed Shovon GIS Analyst Sanjoy Roy Technical Coordinator Mohammad Shahad Mahabub Chowdhury IUCN, International Union for Conservation of Nature Bangladesh Country Office 2015 i The designation of geographical entitles in this book and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of IUCN, International Union for Conservation of Nature concerning the legal status of any country, territory, administration, or concerning the delimitation of its frontiers or boundaries. The biodiversity database and views expressed in this publication are not necessarily reflect those of IUCN, Bangladesh Forest Department and The World Bank. This publication has been made possible because of the funding received from The World Bank through Bangladesh Forest Department to implement the subproject entitled ‘Updating Species Red List of Bangladesh’ under the ‘Strengthening Regional Cooperation for Wildlife Protection (SRCWP)’ Project. Published by: IUCN Bangladesh Country Office Copyright: © 2015 Bangladesh Forest Department and IUCN, International Union for Conservation of Nature and Natural Resources Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holders, provided the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited without prior written permission of the copyright holders. Citation: Of this volume IUCN Bangladesh. 2015. Red List of Bangladesh Volume 2: Mammals. IUCN, International Union for Conservation of Nature, Bangladesh Country Office, Dhaka, Bangladesh, pp. -

North Carolina Department of Agriculture and Consumer Services Veterinary Division
North Carolina Department of Agriculture and Consumer Services Veterinary Division North Carolina Premise Registration Form A complete application should be emailed to [email protected], faxed to (919)733-2277, or mailed to: NC Department of Agriculture Veterinary Division 1030 Mail Service Center Raleigh, NC 27699-1030 If needed, check the following: ☐ Cattle Tags ☐ Swine Tags Premises Owner Account Information Business/Farm Name: Business Type: ☐Individual ☐Incorporated ☐Partnership ☐ LLC ☐ LLP ☐ Government Entity ☐Non-Profit Organization Primary Contact: Phone Number: Mailing Address: City: State: Zip: County: Email Address: Secondary Contact (Optional): Phone Number: Premises Information: Primary location where livestock reside. If animals are managed on separate locations, apply for multiple premises ID’s. Premises Type: ☐Production Unit/Farm/Ranch ☐Market/Collection Point ☐Exhibition ☐Clinic ☐Laboratory ☐ Non-Producer Participant (ie: DHIA, non-animal perm, etc.) ☐Slaughter Plant ☐Other: Premises Name: Premises Address (If different from mailing address): City: State: Zip: County: GPS Coordinates at Entrance (If known): Latitude N Longitude W Species Information: Check all that apply. Quantities of animals are only reported to the state database. This information is protected by GS 106-24.1. This and all other statues can be viewed at www.ncleg.net. If you grow poultry or swine on a contract for a corporation, please indicate production system and corporation for which you grow. Cattle Quantity Equine Quantity Goats Quantity Sheep -

Letter Bill 0..2
HB2554 *LRB10110502SLF55608b* 101ST GENERAL ASSEMBLY State of Illinois 2019 and 2020 HB2554 by Rep. Camille Y. Lilly SYNOPSIS AS INTRODUCED: 720 ILCS 5/48-11 Amends the Criminal Code of 2012. Provides that a person commits unlawful use of an exotic animal in a traveling animal act when he or she knowingly allows for the participation of an exotic animal (rather than an elephant) in a traveling animal act. This offense is a Class A misdemeanor. Defines "exotic animal". LRB101 10502 SLF 55608 b CORRECTIONAL BUDGET AND IMPACT NOTE ACT MAY APPLY A BILL FOR HB2554 LRB101 10502 SLF 55608 b 1 AN ACT concerning criminal law. 2 Be it enacted by the People of the State of Illinois, 3 represented in the General Assembly: 4 Section 5. The Criminal Code of 2012 is amended by changing 5 Section 48-11 as follows: 6 (720 ILCS 5/48-11) 7 Sec. 48-11. Unlawful use of an exotic animal elephant in a 8 traveling animal act. 9 (a) Definitions. As used in this Section: 10 "Exotic animal" means any animal that is native to a 11 foreign country or of foreign origin or character, is not 12 native to the United States, or was introduced from abroad 13 including, but not limited to, lions, tigers, leopards, 14 elephants, camels, antelope, anteaters, kangaroos, and water 15 buffalo and species of foreign domestic cattle, such as Ankole, 16 Gayal, and Yak or a wild animal. 17 "Mobile or traveling animal housing facility" means a 18 transporting vehicle such as a truck, trailer, or railway car 19 used to transport or house animals while traveling to an 20 exhibition or other performance. -

Hippotragus Equinus – Roan Antelope
Hippotragus equinus – Roan Antelope authorities as there may be no significant genetic differences between the two. Many of the Roan Antelope in South Africa are H. e. cottoni or equinus x cottoni (especially on private properties). Assessment Rationale This charismatic antelope exists at low density within the assessment region, occurring in savannah woodlands and grasslands. Currently (2013–2014), there are an observed 333 individuals (210–233 mature) existing on nine formally protected areas within the natural distribution range. Adding privately protected subpopulations and an Cliff & Suretha Dorse estimated 0.8–5% of individuals on wildlife ranches that may be considered wild and free-roaming, yields a total mature population of 218–294 individuals. Most private Regional Red List status (2016) Endangered subpopulations are intensively bred and/or kept in camps C2a(i)+D*†‡ to exclude predators and to facilitate healthcare. Field National Red List status (2004) Vulnerable D1 surveys are required to identify potentially eligible subpopulations that can be included in this assessment. Reasons for change Non-genuine: While there was an historical crash in Kruger National Park New information (KNP) of 90% between 1986 and 1993, the subpopulation Global Red List status (2008) Least Concern has since stabilised at c. 50 individuals. Overall, over the past three generations (1990–2015), based on available TOPS listing (NEMBA) Vulnerable data for nine formally protected areas, there has been a CITES listing None net population reduction of c. 23%, which indicates an ongoing decline but not as severe as the historical Endemic Edge of Range reduction. Further long-term data are needed to more *Watch-list Data †Watch-list Threat ‡Conservation Dependent accurately estimate the national population trend. -
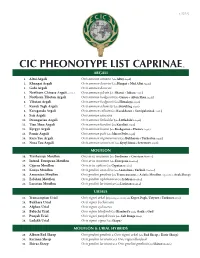
Cic Pheonotype List Caprinae©
v. 5.25.12 CIC PHEONOTYPE LIST CAPRINAE © ARGALI 1. Altai Argali Ovis ammon ammon (aka Altay Argali) 2. Khangai Argali Ovis ammon darwini (aka Hangai & Mid Altai Argali) 3. Gobi Argali Ovis ammon darwini 4. Northern Chinese Argali - extinct Ovis ammon jubata (aka Shansi & Jubata Argali) 5. Northern Tibetan Argali Ovis ammon hodgsonii (aka Gansu & Altun Shan Argali) 6. Tibetan Argali Ovis ammon hodgsonii (aka Himalaya Argali) 7. Kuruk Tagh Argali Ovis ammon adametzi (aka Kuruktag Argali) 8. Karaganda Argali Ovis ammon collium (aka Kazakhstan & Semipalatinsk Argali) 9. Sair Argali Ovis ammon sairensis 10. Dzungarian Argali Ovis ammon littledalei (aka Littledale’s Argali) 11. Tian Shan Argali Ovis ammon karelini (aka Karelini Argali) 12. Kyrgyz Argali Ovis ammon humei (aka Kashgarian & Hume’s Argali) 13. Pamir Argali Ovis ammon polii (aka Marco Polo Argali) 14. Kara Tau Argali Ovis ammon nigrimontana (aka Bukharan & Turkestan Argali) 15. Nura Tau Argali Ovis ammon severtzovi (aka Kyzyl Kum & Severtzov Argali) MOUFLON 16. Tyrrhenian Mouflon Ovis aries musimon (aka Sardinian & Corsican Mouflon) 17. Introd. European Mouflon Ovis aries musimon (aka European Mouflon) 18. Cyprus Mouflon Ovis aries ophion (aka Cyprian Mouflon) 19. Konya Mouflon Ovis gmelini anatolica (aka Anatolian & Turkish Mouflon) 20. Armenian Mouflon Ovis gmelini gmelinii (aka Transcaucasus or Asiatic Mouflon, regionally as Arak Sheep) 21. Esfahan Mouflon Ovis gmelini isphahanica (aka Isfahan Mouflon) 22. Larestan Mouflon Ovis gmelini laristanica (aka Laristan Mouflon) URIALS 23. Transcaspian Urial Ovis vignei arkal (Depending on locality aka Kopet Dagh, Ustyurt & Turkmen Urial) 24. Bukhara Urial Ovis vignei bocharensis 25. Afghan Urial Ovis vignei cycloceros 26.