Zoologische Mededelingen
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies
www.nature.com/scientificreports OPEN Kanyawara Virus: A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies Received: 19 April 2017 Accepted: 25 May 2017 Infesting Previously Unknown Published: xx xx xxxx Pteropodid Bats in Uganda Tony L. Goldberg 1,2,3, Andrew J. Bennett1, Robert Kityo3, Jens H. Kuhn4 & Colin A. Chapman3,5 Bats are natural reservoir hosts of highly virulent pathogens such as Marburg virus, Nipah virus, and SARS coronavirus. However, little is known about the role of bat ectoparasites in transmitting and maintaining such viruses. The intricate relationship between bats and their ectoparasites suggests that ectoparasites might serve as viral vectors, but evidence to date is scant. Bat flies, in particular, are highly specialized obligate hematophagous ectoparasites that incidentally bite humans. Using next- generation sequencing, we discovered a novel ledantevirus (mononegaviral family Rhabdoviridae, genus Ledantevirus) in nycteribiid bat flies infesting pteropodid bats in western Uganda. Mitochondrial DNA analyses revealed that both the bat flies and their bat hosts belong to putative new species. The coding-complete genome of the new virus, named Kanyawara virus (KYAV), is only distantly related to that of its closest known relative, Mount Elgon bat virus, and was found at high titers in bat flies but not in blood or on mucosal surfaces of host bats. Viral genome analysis indicates unusually low CpG dinucleotide depletion in KYAV compared to other ledanteviruses and rhabdovirus groups, with KYAV displaying values similar to rhabdoviruses of arthropods. Our findings highlight the possibility of a yet- to-be-discovered diversity of potentially pathogenic viruses in bat ectoparasites. Bats (order Chiroptera) represent the second largest order of mammals after rodents (order Rodentia). -
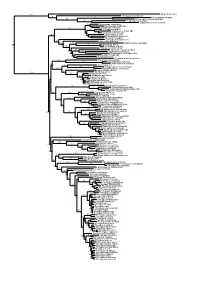
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

Final Report on the Project
BP Conservation Programme (CLP) 2005 PROJECT NO. 101405 - BRONZE AWARD WINNER ECOLOGY, DISTRIBUTION, STATUS AND PROTECTION OF THREE CONGOLESE FRUIT BATS FINAL REPORT Patrick KIPALU Team Leader Observatoire Congolais pour la Protection de l’Environnement OCPE – ong Kinshasa – Democratic Republic of the Congo E-mail: [email protected] APRIL 2009 1 Table of Content Acknowledgements…………………………………………………………………. p3 I. Project Summary……………………………………………………………….. p4 II. Introduction…………………………………………………………………… p4-p7 III. Materials and Methods ……………………………………………………….. p7-p10 IV. Results per Study Site…………………………………………………………. p10-p15 1. Pointe-Noire ………………………………………………………….. p10-p12 2. Mayumbe Forest /Luki Reserve……………………………………….. p12-p13 3. Zongo Forest…………………………………………………………... p14 4. Mbanza-Ngungu ………………………………………………………. P15 V. General Results ………………………………………………………………p15-p16 VI. Discussions……………………………………………………………………p17-18 VII. Conclusion and Recommendations……………………………………….p 18-p19 VIII. Bibliography………………………………………………………………p20-p21 Acknowledgements 2 The OCPE (Observatoire Congolais pour la Protection de l’Environnement) project team would like to start by expressing our gratefulness and saying thank you to the BP Conservation Program, which has funded the execution of this project. The OCPE also thanks the Van Tienhoven Foundation which provided a further financial support. Without these organisations, execution of the project would not have been possible. We would like to thank specially the BPCP “dream team”: Marianne D. Carter, Robyn Dalzen and our regretted Kate Stoke for their time, advices, expertise and care, which helped us to complete this work, Our special gratitude goes to Dr. Wim Bergmans, who was the hero behind the scene from the conception to the execution of the research work. Without his expertise, advices and network it would had been difficult for the project team to produce any result from this project. -

Index of Handbook of the Mammals of the World. Vol. 9. Bats
Index of Handbook of the Mammals of the World. Vol. 9. Bats A agnella, Kerivoula 901 Anchieta’s Bat 814 aquilus, Glischropus 763 Aba Leaf-nosed Bat 247 aladdin, Pipistrellus pipistrellus 771 Anchieta’s Broad-faced Fruit Bat 94 aquilus, Platyrrhinus 567 Aba Roundleaf Bat 247 alascensis, Myotis lucifugus 927 Anchieta’s Pipistrelle 814 Arabian Barbastelle 861 abae, Hipposideros 247 alaschanicus, Hypsugo 810 anchietae, Plerotes 94 Arabian Horseshoe Bat 296 abae, Rhinolophus fumigatus 290 Alashanian Pipistrelle 810 ancricola, Myotis 957 Arabian Mouse-tailed Bat 164, 170, 176 abbotti, Myotis hasseltii 970 alba, Ectophylla 466, 480, 569 Andaman Horseshoe Bat 314 Arabian Pipistrelle 810 abditum, Megaderma spasma 191 albatus, Myopterus daubentonii 663 Andaman Intermediate Horseshoe Arabian Trident Bat 229 Abo Bat 725, 832 Alberico’s Broad-nosed Bat 565 Bat 321 Arabian Trident Leaf-nosed Bat 229 Abo Butterfly Bat 725, 832 albericoi, Platyrrhinus 565 andamanensis, Rhinolophus 321 arabica, Asellia 229 abramus, Pipistrellus 777 albescens, Myotis 940 Andean Fruit Bat 547 arabicus, Hypsugo 810 abrasus, Cynomops 604, 640 albicollis, Megaerops 64 Andersen’s Bare-backed Fruit Bat 109 arabicus, Rousettus aegyptiacus 87 Abruzzi’s Wrinkle-lipped Bat 645 albipinnis, Taphozous longimanus 353 Andersen’s Flying Fox 158 arabium, Rhinopoma cystops 176 Abyssinian Horseshoe Bat 290 albiventer, Nyctimene 36, 118 Andersen’s Fruit-eating Bat 578 Arafura Large-footed Bat 969 Acerodon albiventris, Noctilio 405, 411 Andersen’s Leaf-nosed Bat 254 Arata Yellow-shouldered Bat 543 Sulawesi 134 albofuscus, Scotoecus 762 Andersen’s Little Fruit-eating Bat 578 Arata-Thomas Yellow-shouldered Talaud 134 alboguttata, Glauconycteris 833 Andersen’s Naked-backed Fruit Bat 109 Bat 543 Acerodon 134 albus, Diclidurus 339, 367 Andersen’s Roundleaf Bat 254 aratathomasi, Sturnira 543 Acerodon mackloti (see A. -

High Diversity of West African Bat Malaria Parasites and a Tight Link with Rodent Plasmodium Taxa
High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa Juliane Schaera,b,1,2, Susan L. Perkinsc,1,2, Jan Decherd, Fabian H. Leendertze, Jakob Fahrf,g, Natalie Weberh, and Kai Matuschewskia,i aParasitology Unit, Max Planck Institute for Infection Biology, 10117 Berlin, Germany; bMuseum für Naturkunde, Leibniz Institute for Research on Evolution and Biodiversity, 10115 Berlin, Germany; cSackler Institute for Comparative Genomics, American Museum of Natural History, New York, NY 10024; dSection of Mammals, Zoologisches Forschungsmuseum A. König, 53113 Bonn, Germany; eRobert Koch Institute, 13302 Berlin, Germany; fZoological Institute, Technische Universität Braunschweig, 38106 Braunschweig, Germany; gDepartment of Migration and Immuno-ecology, Max Planck Institute for Ornithology, 78315 Radolfzell, Germany; hInstitute of Experimental Ecology, University of Ulm, 89069 Ulm, Germany; and iInstitute of Biology, Humboldt University, 10117 Berlin, Germany Edited by Jitender P. Dubey, US Department of Agriculture, Beltsville, MD, and approved September 10, 2013 (received for review June 10, 2013) As the only volant mammals, bats are captivating for their high to the late 19th century (10). The corresponding vectors for most taxonomic diversity, for their vital roles in ecosystems—particularly as bat parasites remain unknown. Similarly, the phylogenetic rela- pollinators and insectivores—and, more recently, for their important tionships for the majority of these parasites remain enigmatic. roles in the maintenance and transmission of zoonotic viral diseases. Here, we present a unique systematic analysis of Haemosporida Genome sequences have identified evidence for a striking expansion in a diverse species assemblage of bats. For this study, we per- of and positive selection in gene families associated with immunity. -

Interrogating Phylogenetic Discordance Resolves Deep Splits
Interrogating Phylogenetic Discordance Resolves Deep Splits in the Rapid Radiation of Old World Fruit Bats (Chiroptera: Pteropodidae) Nicolas Nesi, Georgia Tsagkogeorga, Susan Tsang, Violaine Nicolas, Aude Lalis, Annette Scanlon, Silke Riesle-Sbarbaro, Sigit Wiantoro, Alan Hitch, Javier Juste, et al. To cite this version: Nicolas Nesi, Georgia Tsagkogeorga, Susan Tsang, Violaine Nicolas, Aude Lalis, et al.. Interrogat- ing Phylogenetic Discordance Resolves Deep Splits in the Rapid Radiation of Old World Fruit Bats (Chiroptera: Pteropodidae). Systematic Biology, Oxford University Press (OUP), 2021, 10.1093/sys- bio/syab013. hal-03167896 HAL Id: hal-03167896 https://hal.sorbonne-universite.fr/hal-03167896 Submitted on 12 Mar 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Interrogating Phylogenetic Discordance Resolves Deep Splits in the Rapid Radiation of Old World Fruit Bats (Chiroptera: Pteropodidae) Downloaded from https://academic.oup.com/sysbio/advance-article/doi/10.1093/sysbio/syab013/6161783 by Sorbonne Université user on 12 March 2021 NICOLAS NESI1, GEORGIA TSAGKOGEORGA1, SUSAN M. TSANG2,3, VIOLAINE NICOLAS4, AUDE LALIS4, ANNETTE T. SCANLON5, SILKE A. RIESLE- SBARBARO6,7†, SIGIT WIANTORO8, ALAN T. HITCH9, JAVIER JUSTE10, CORINNA A. PINZARI11, FRANK J. BONACCORSO12, CHRISTOPHER M. -

Cryptosporidium Spp. in Wild and Captive Australian Flying Foxes
Cryptosporidium spp. in wild and captive Australian flying foxes (genus: Pteropus) Sabine Eva Schiller, Bachelor of Philosophy Macquarie University, North Ryde, NSW, 2109, Australia Department of Biological Sciences Tel.: 61 2 9850 9259; Fax: 61 2 9850 8245 Submission date: 09.10.2015 1 Table of Contents Title Page……………………………………………………………………………………………………………………………………….….1 Table of Contents……………………………………………………………………………………………………………………………2-3 Abstract………………………………………………………………………………………………………………..……………………………4 Declaration………………………………………………………………………………………………………………………………………..5 Acknowledgements……………………………………………………………………………………………………………………………6 Abbreviations…………………………………………………………………………………………………………………………………….7 1. Introduction .................................................................................................................................... 8 1.1. Biology of Cryptosporidium .................................................................................................... 9 1.1.1. Cryptosporidium taxonomy and species identification....................................................... 9 1.1.2. Cryptosporidium: host specificity and cryptosporidiosis .................................................. 10 1.1.3. Cryptosporidium lifecycle .................................................................................................. 11 1.1.4. Sources and transmission pathways ................................................................................. 14 1.2. Zooanthroponotic pathogen transmission in wildlife and livestock .................................... -

Metagenomic Snapshots of Viral Components in Guinean Bats
microorganisms Communication Metagenomic Snapshots of Viral Components in Guinean Bats Roberto J. Hermida Lorenzo 1,†,Dániel Cadar 2,† , Fara Raymond Koundouno 3, Javier Juste 4,5 , Alexandra Bialonski 2, Heike Baum 2, Juan Luis García-Mudarra 4, Henry Hakamaki 2, András Bencsik 2, Emily V. Nelson 2, Miles W. Carroll 6,7, N’Faly Magassouba 3, Stephan Günther 2,8, Jonas Schmidt-Chanasit 2,9 , César Muñoz Fontela 2,8 and Beatriz Escudero-Pérez 2,8,* 1 Morcegos de Galicia, Magdalena G-2, 2o izq, 15320 As Pontes de García Rodríguez (A Coruña), Spain; [email protected] 2 WHO Collaborating Centre for Arbovirus and Haemorrhagic Fever Reference and Research, Bernhard Nocht Institute for Tropical Medicine, 20359 Hamburg, Germany; [email protected] (D.C.); [email protected] (A.B.); [email protected] (H.B.); [email protected] (H.H.); [email protected] (A.B.); [email protected] (E.V.N.); [email protected] (S.G.); [email protected] (J.S.-C.); [email protected] (C.M.F.) 3 Laboratoire des Fièvres Hémorragiques en Guinée, Université Gamal Abdel Nasser de Conakry, Commune de Matoto, Conakry, Guinea; [email protected] (F.R.K.); [email protected] (N.M.) 4 Estación Biológica de Doñana, CSIC, 41092 Seville, Spain; [email protected] (J.J.); [email protected] (J.L.G.-M.) 5 CIBER Epidemiology and Public Health (CIBERESP), 28029 Madrid, Spain 6 Public Health England, Porton Down, Wiltshire SP4 0JG, UK; [email protected] 7 Wellcome Centre for Human Genetics, Nuffield Department of Medicine, Oxford University, Oxford OX3 7BN, UK Citation: Hermida Lorenzo, R.J.; 8 German Centre for Infection Research (DZIF), Partner Site Hamburg-Luebeck-Borstel, Cadar, D.; Koundouno, F.R.; Juste, J.; 38124 Braunschweig, Germany Bialonski, A.; Baum, H.; 9 Faculty of Mathematics, Informatics and Natural Sciences, Universität Hamburg, 20148 Hamburg, Germany García-Mudarra, J.L.; Hakamaki, H.; * Correspondence: [email protected] Bencsik, A.; Nelson, E.V.; et al. -

Master's Thesis
MASTER’S THESIS ‘Megabats’ Stephen Turnbull Department of Biological Sciences Faculty of Science Aarhus University, Denmark [email protected] Supervisor: Associate Professor Jens M. Olesen Cover photograph: Dorte Nyhagen Introduction Why megabats? A brief explanation of my experiences with megabats. I first came across megabats when studying for my honours project at Aberdeen University under the supervision of Professor Paul Racey – an intimidating yet extremely likeable giant of the bat world. I was to study Pteropus rodricensis, the famed golden fruit bat, endemic to the island of Rodrigues; a tiny far-flung speck in the Indian Ocean, politically aligned with Mauritius. I had some idea of what to expect, but no firm plans of how to carry out my studies, relying instead on my confident ability to improvise. Upon arrival, the island presented itself as a catalogue of environmental short-sightedness and ecological collapse, yet the fruit bats clung on to their perilous existence, saved from extinction by the irregular topography of some parts of the island. In a near- vertical and densely vegetated gorge, the bats could roost in peace during the day, flying to their feeding sites each evening at dusk, their destinations presumably carefully planned the previous night. I quickly came to realise a number of problems inherently linked with the study of fruit-bats. Firstly, they’re nocturnal. This presents a whole host of difficulties, not least of which being the absence of daylight. Secondly, there was no way in which to access their roost site, and even if I could, my clumsy approach would disturb them. -

Observations, Discussions and Updates
African Bat Conservation News 2 May 2017 vol. 45 ISSN 1812-1268 Observations, Discussions and Updates RECENT CHANGES IN AFRicAN BAT TAXONOMY (2015 – 2017) VICTOR VAN CAKENBERGHE1,2 AND ERNest C.J. SEAMARK2,3 1 University of Antwerp, Department of Biology, Lab for Functional Morphology, Campus Drie Eiken, Universiteitsplein, 1, B-2610 Antwerpen (Wilrijk), Belgium. 2 AfricanBats NPC, 357 Botha Ave, Kloofsig, 0157. 3 Centre for Wildlife Management, University of Pretoria, Private Bag X20 Hatfield, Pretoria 0028, South Africa. †NECROMANTIDAE Sigé, 2011 was discovered. The specific epithet is given in honour of Mary Knight, Managing Editor of the American Museum of Natural History †Necromantis Weithofer, 1887 Scientific Publications, in recognition of the enormous contributions she has made to dissemination of the results of scientific research †Necromantis fragmentum Ravel, in Ravel, Adaci, over the years, as well as her lifelong devotion to the people and Bensalah, Charruault, Essid, AMmar, Marzougui, culture of Egypt. Mahboubi, Mebrouk, Merzeraud, Vianey-Liaud, Tabuce and Marivaux, 2016 Eidolinae Almeida, Giannini and Simmons, 2016 Locality and Horizon: Chambi (CBI) loci 1, 2 and 3, late lower This subfamily was created by ALMEIDA et al. (2016: 84) to Eocene - middle Eocene, Djebel Chambi, situated in the Kasserine distinguish monophyletic groups within the Pteropodidae. region, Tunisia. Currently recognized genera of Eidolinae: Eidolon Rafinesque, Etymology: From the Latin ‘fragmentum’, meaning fragment 1815. or debris, referring to the fragmentary character of the material attributed to this taxon. Pteropodinae Gray 1821 Subfamily arrangement based on ALMEIDA et al. (2016: 83). †AEGYPTONYCTERIDAE Simmons, Seiffert and Gunnell, Currently recognized genera of Pteropodinae: Acerodon Jourdan, 2016 1837, Macroglossus F. -

Exploitation of Bats for Bushmeat and Medicine
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Springer - Publisher Connector Chapter 12 Exploitation of Bats for Bushmeat and Medicine Tammy Mildenstein, Iroro Tanshi and Paul A. Racey Abstract Bat hunting for consumption as bushmeat and medicine is widespread and affects at least 167 species of bats (or c. 13 % of the world’s bat species), in Africa, Asia, across the islands of Oceania, and to a lesser extent in Central and South America. Hunting is particularly prevalent among the large-bodied fruit bats of the Old World tropics, where half (50 %, 92/183) the extant species in the family Pteropodidae are hunted. Pteropodids that are hunted are six times more likely to be Red Listed as threatened: 66 % of species in IUCN threatened categories (CR, EN, VU, NT), compared to 11 % of species in the ‘Least Concern’ (LC) category. However, there still appears to be an information gap at the international level. One third of the hunted species on the Red List are not considered threatened by that hunting, and nearly a quarter of the bat species included in this review are not listed as hunted in IUCN Red List species accounts. This review has resulted in a com- prehensive list of hunted bats that doubles the number of species known from either the IUCN Red List species accounts or a questionnaire circulated in 2004. More research is needed on the impacts of unregulated hunting, as well as on the sustain- ability of regulated hunting programs. In the absence of population size and growth data, legislators and managers should be precautionary in their attitude towards T. -

A Phylogenetic Tree for Pteropus and Related Genera (Chiroptera: Pteropodidae) ⇑ Francisca C
Molecular Phylogenetics and Evolution 77 (2014) 83–95 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Each flying fox on its own branch: A phylogenetic tree for Pteropus and related genera (Chiroptera: Pteropodidae) ⇑ Francisca C. Almeida a, ,1, Norberto P. Giannini a,b, Nancy B. Simmons a, Kristofer M. Helgen c a American Museum of Natural History, Division of Vertebrate Zoology, Department of Mammalogy, Central Park West at 79th Street, New York, NY, United States b Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Universidad Nacional de Tucumán, Faculdad de Ciencias Naturales e Instituto Miguel Lillo, Miguel Lillo, 205, San Miguel de Tucumán 4000, Argentina c Division of Mammals, National Museum of Natural History, Smithsonian Institution, Washington, D.C. 20013-7012, United States article info abstract Article history: Pteropodidae is a diverse Old World family of non-echolocating, frugivorous and nectarivorous bats that Received 20 December 2013 includes the flying foxes (genus Pteropus) and allied genera. The subfamily Pteropodinae includes the Revised 9 March 2014 largest living bats and is distributed across an immense geographic range from islands in East Africa to Accepted 11 March 2014 the Cook Islands of Polynesia. These bats are keystone species in their ecosystems and some carry zoo- Available online 22 March 2014 notic diseases that are increasingly a focus of interest in biomedical research. Here we present a compre- hensive phylogeny for pteropodines focused on Pteropus. The analyses included 50 of the 63 species of Keywords: Pteropus and 11 species from 7 related genera. We obtained sequences of the cytochrome b and the 12S Pteropus rRNA mitochondrial genes for all species and sequences of the nuclear RAG1, vWF, and BRCA1 genes for a Pteropodidae Chiroptera subsample of taxa.