Comparative Phylogeography of African Fruit Bats
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies
www.nature.com/scientificreports OPEN Kanyawara Virus: A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies Received: 19 April 2017 Accepted: 25 May 2017 Infesting Previously Unknown Published: xx xx xxxx Pteropodid Bats in Uganda Tony L. Goldberg 1,2,3, Andrew J. Bennett1, Robert Kityo3, Jens H. Kuhn4 & Colin A. Chapman3,5 Bats are natural reservoir hosts of highly virulent pathogens such as Marburg virus, Nipah virus, and SARS coronavirus. However, little is known about the role of bat ectoparasites in transmitting and maintaining such viruses. The intricate relationship between bats and their ectoparasites suggests that ectoparasites might serve as viral vectors, but evidence to date is scant. Bat flies, in particular, are highly specialized obligate hematophagous ectoparasites that incidentally bite humans. Using next- generation sequencing, we discovered a novel ledantevirus (mononegaviral family Rhabdoviridae, genus Ledantevirus) in nycteribiid bat flies infesting pteropodid bats in western Uganda. Mitochondrial DNA analyses revealed that both the bat flies and their bat hosts belong to putative new species. The coding-complete genome of the new virus, named Kanyawara virus (KYAV), is only distantly related to that of its closest known relative, Mount Elgon bat virus, and was found at high titers in bat flies but not in blood or on mucosal surfaces of host bats. Viral genome analysis indicates unusually low CpG dinucleotide depletion in KYAV compared to other ledanteviruses and rhabdovirus groups, with KYAV displaying values similar to rhabdoviruses of arthropods. Our findings highlight the possibility of a yet- to-be-discovered diversity of potentially pathogenic viruses in bat ectoparasites. Bats (order Chiroptera) represent the second largest order of mammals after rodents (order Rodentia). -

Checklist of the Mammals of Indonesia
CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation i ii CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation By Ibnu Maryanto Maharadatunkamsi Anang Setiawan Achmadi Sigit Wiantoro Eko Sulistyadi Masaaki Yoneda Agustinus Suyanto Jito Sugardjito RESEARCH CENTER FOR BIOLOGY INDONESIAN INSTITUTE OF SCIENCES (LIPI) iii © 2019 RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI) Cataloging in Publication Data. CHECKLIST OF THE MAMMALS OF INDONESIA: Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation/ Ibnu Maryanto, Maharadatunkamsi, Anang Setiawan Achmadi, Sigit Wiantoro, Eko Sulistyadi, Masaaki Yoneda, Agustinus Suyanto, & Jito Sugardjito. ix+ 66 pp; 21 x 29,7 cm ISBN: 978-979-579-108-9 1. Checklist of mammals 2. Indonesia Cover Desain : Eko Harsono Photo : I. Maryanto Third Edition : December 2019 Published by: RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI). Jl Raya Jakarta-Bogor, Km 46, Cibinong, Bogor, Jawa Barat 16911 Telp: 021-87907604/87907636; Fax: 021-87907612 Email: [email protected] . iv PREFACE TO THIRD EDITION This book is a third edition of checklist of the Mammals of Indonesia. The new edition provides remarkable information in several ways compare to the first and second editions, the remarks column contain the abbreviation of the specific island distributions, synonym and specific location. Thus, in this edition we are also corrected the distribution of some species including some new additional species in accordance with the discovery of new species in Indonesia. -

New Records of the Togo Toad, Sclerophrys Togoensis, from South-Eastern Ivory Coast
Herpetology Notes, volume 12: 501-508 (2019) (published online on 19 May 2019) New records of the Togo Toad, Sclerophrys togoensis, from south-eastern Ivory Coast Basseu Aude-Inès Gongomin1, N’Goran Germain Kouamé1,*, and Mark-Oliver Rödel2 Abstract. Reported are new records of the forest toad, Sclerophrys togoensis, from south-eastern Ivory Coast. A small population was found in the rainforest of Mabi and Yaya Classified Forests. These forests and Taï National Park in the western part of the country are the only known and remaining Ivorian habitats of this species. Sclerophrys togoensis is confined to primary and slightly degraded rainforest. Known sites should be urgently and effectively protected from further forest loss. Keywords. Amphibia, Anura, Bufonidae, Conservation, Distribution, Mabi/Yaya Classified Forests, Upper Guinea forest Introduction In Ivory Coast the known records of S. togoensis are from the Cavally and Haute Dodo Classified Forests The toad Sclerophrys togoensis (Ahl, 1924) has been (Rödel and Branch, 2002), and the Taï National Park described from Bismarckburg in Togo (Ahl, 1924). Apart and its surroundings (e.g. Ernst and Rödel, 2006; Hillers from a parasitological study (Bourgat, 1978), no recent et al., 2008), all situated in the westernmost part of the records are known from that country (Ségniagbeto et al., country (Fig. 1). During a decade of conflict, both 2007; Hillers et al., 2009). Further records have been classified forests have been deforested (P.J. Adeba, pers. published from southern Ghana (Kouamé et al., 2007; comm.), thus restricting the species known Ivorian range Hillers et al., 2009), western Ivory Coast (e.g. -

Bat Distribution Size Or Shape As Determinant of Viral Richness in African Bats
Bat Distribution Size or Shape as Determinant of Viral Richness in African Bats Gae¨l D. Maganga1,2., Mathieu Bourgarel1,3,4*., Peter Vallo5,6, Thierno D. Dallo7, Carine Ngoagouni8, Jan Felix Drexler7, Christian Drosten7, Emmanuel R. Nakoune´ 8, Eric M. Leroy1,9, Serge Morand3,10,11. 1 Centre International de Recherches Me´dicales de Franceville, Franceville, Gabon, 2 Institut National Supe´rieur d’Agronomie et de Biotechnologies (INSAB), Franceville, Gabon, 3 CIRAD, UPR AGIRs, Montpellier, France, 4 CIRAD, UPR AGIRs, Harare, Zimbabwe, 5 Institute of Vertebrate Biology, Academy of Sciences of the Czech Republic, Brno, Czech Republic, 6 Institute of Experimental Ecology, Ulm University, Ulm, Germany, 7 Institute of Virology, University of Bonn Medical Centre, Bonn, Germany, 8 Institut Pasteur de Bangui, Bangui, Re´publique Centrafricaine, 9 Institut de Recherche pour le De´veloppement, UMR 224 (MIVEGEC), IRD/CNRS/UM1, Montpellier, France, 10 Institut des Sciences de l’Evolution, CNRS-UM2, CC065, Universite´ de Montpellier 2, Montpellier, France, 11 Centre d’Infectiologie Christophe Me´rieux du Laos, Vientiane, Lao PDR Abstract The rising incidence of emerging infectious diseases (EID) is mostly linked to biodiversity loss, changes in habitat use and increasing habitat fragmentation. Bats are linked to a growing number of EID but few studies have explored the factors of viral richness in bats. These may have implications for role of bats as potential reservoirs. We investigated the determinants of viral richness in 15 species of African bats (8 Pteropodidae and 7 microchiroptera) in Central and West Africa for which we provide new information on virus infection and bat phylogeny. -

Mammals of Jordan
© Biologiezentrum Linz/Austria; download unter www.biologiezentrum.at Mammals of Jordan Z. AMR, M. ABU BAKER & L. RIFAI Abstract: A total of 78 species of mammals belonging to seven orders (Insectivora, Chiroptera, Carni- vora, Hyracoidea, Artiodactyla, Lagomorpha and Rodentia) have been recorded from Jordan. Bats and rodents represent the highest diversity of recorded species. Notes on systematics and ecology for the re- corded species were given. Key words: Mammals, Jordan, ecology, systematics, zoogeography, arid environment. Introduction In this account we list the surviving mammals of Jordan, including some reintro- The mammalian diversity of Jordan is duced species. remarkable considering its location at the meeting point of three different faunal ele- Table 1: Summary to the mammalian taxa occurring ments; the African, Oriental and Palaearc- in Jordan tic. This diversity is a combination of these Order No. of Families No. of Species elements in addition to the occurrence of Insectivora 2 5 few endemic forms. Jordan's location result- Chiroptera 8 24 ed in a huge faunal diversity compared to Carnivora 5 16 the surrounding countries. It shelters a huge Hyracoidea >1 1 assembly of mammals of different zoogeo- Artiodactyla 2 5 graphical affinities. Most remarkably, Jordan Lagomorpha 1 1 represents biogeographic boundaries for the Rodentia 7 26 extreme distribution limit of several African Total 26 78 (e.g. Procavia capensis and Rousettus aegypti- acus) and Palaearctic mammals (e. g. Eri- Order Insectivora naceus concolor, Sciurus anomalus, Apodemus Order Insectivora contains the most mystacinus, Lutra lutra and Meles meles). primitive placental mammals. A pointed snout and a small brain case characterises Our knowledge on the diversity and members of this order. -

Dahomey Gap” - Une Contribution À L’Histoire De La Végétation Au Sud-Bénin Et Sud-Ouest Du Nigeria
Le projet ”Dahomey Gap” - une contribution à l’histoire de la végétation au Sud-Bénin et Sud-ouest du Nigeria. Aziz Ballouche, Akpovi Akoegninou, Katharina Neumann, Ulrich Salzmann, Adebisi Sowunmi To cite this version: Aziz Ballouche, Akpovi Akoegninou, Katharina Neumann, Ulrich Salzmann, Adebisi Sowunmi. Le projet ”Dahomey Gap” - une contribution à l’histoire de la végétation au Sud-Bénin et Sud-ouest du Nigeria.. Berichte des Frankfurter Sonderforschungsbereiches 268, 2000, 14, pp.237-251. halshs- 00130147 HAL Id: halshs-00130147 https://halshs.archives-ouvertes.fr/halshs-00130147 Submitted on 12 Feb 2007 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Berichte des Sonderforschungsbereichs 268, Band 14, Frankfurt a.M. 2000: 237-251 LE PROJET "DAHOMEY GAP": UNE CONTRIBUTION A L'HISTOIRE DE LA VEGETATION AU SUD-BENIN ET SUD- OUEST DU NIGERIA Aziz Ballouche, Akpovi Akoègninou, Katharina Neumann, Ulrich Salzmann et M. Adebisi Sowunmi Résumé La coupure que l'on constate dans les forêts denses africaines au niveau du Togo et du Bénin soulève de nombreuses questions sur les rapports forêt/savane au cours des derniers millénaires. Le projet Dahomey Gap vise à y répondre, par une approche pluridisciplinaire intégrant botanique et histoire de la végétation dans le Sud-Bénin et le Sud-Ouest du Nigéria. -

Zoologische Mededelingen
ZOOLOGISCHE MEDEDELINGEN UITGEGEVEN DOOR HET RIJKSMUSEUM VAN NATUURLIJKE HISTORIE TE LEIDEN (MINISTERIE VAN CULTUUR, RECREATIE EN MAATSCHAPPELIJK WERK) Deel 55 no. 14 4 maart 1980 A NEW FRUIT BAT OF THE GENUS MYONYCTERIS MATSCHIE, 1899, FROM EASTERN KENYA AND TANZANIA (MAMMALIA, MEGACHIROPTERA) by W. BERGMANS Instituut voor Taxonomische Zoölogie, Universiteit van Amsterdam With 4 text-figures ABSTRACT Myonycteris relicta n. sp. is described from the Shimba Hills in southeast Kenya and from the Usambara Mountains in northeast Tanzania. The species is larger than the only other known African mainland species of the genus, Myonycteris torquata (Dobson, 1878), from the Central and West African rain forests and, if compared to M. torquata and the only other species in the genus, M. brachycephala (Bocage, 1889) from São Tomé, has a relatively longer rostrum, a more deflected cranial axis, and further differs in number, shape and position of its teeth. The new species provides new arguments for the relationship between the genera Myonycteris Matschie, 1899, and Lissonycteris Andersen, 1912. It is believed that Myonycteris relicta may be a forest species and as such restricted to isolated East African forests. INTRODUCTION During a visit to the Zoologisches Museum in Berlin (ZMB), in April 1979, the author found two fruit bat specimens from the Tanzanian Usa- mbara Mountains, which proved to represent an undescribed taxon. Later, in June 1979, Dr C. Smeenk of the Rijksmuseum van Natuurlijke Historie at Leiden (RMNH) recognized a third specimen of this taxon in newly acquired material from the Shimba Hills in southeast Kenya. The bats differ on specific level from all other known fruit bats, and are described in the present paper. -

Chiroptera: Pteropodidae)
Chapter 6 Phylogenetic Relationships of Harpyionycterine Megabats (Chiroptera: Pteropodidae) NORBERTO P. GIANNINI1,2, FRANCISCA CUNHA ALMEIDA1,3, AND NANCY B. SIMMONS1 ABSTRACT After almost 70 years of stability following publication of Andersen’s (1912) monograph on the group, the systematics of megachiropteran bats (Chiroptera: Pteropodidae) was thrown into flux with the advent of molecular phylogenetics in the 1980s—a state where it has remained ever since. One particularly problematic group has been the Austromalayan Harpyionycterinae, currently thought to include Dobsonia and Harpyionycteris, and probably also Aproteles.Inthis contribution we revisit the systematics of harpyionycterines. We examine historical hypotheses of relationships including the suggestion by O. Thomas (1896) that the rousettine Boneia bidens may be related to Harpyionycteris, and report the results of a series of phylogenetic analyses based on new as well as previously published sequence data from the genes RAG1, RAG2, vWF, c-mos, cytb, 12S, tVal, 16S,andND2. Despite a striking lack of morphological synapomorphies, results of our combined analyses indicate that Boneia groups with Aproteles, Dobsonia, and Harpyionycteris in a well-supported, expanded Harpyionycterinae. While monophyly of this group is well supported, topological changes within this clade across analyses of different data partitions indicate conflicting phylogenetic signals in the mitochondrial partition. The position of the harpyionycterine clade within the megachiropteran tree remains somewhat uncertain. Nevertheless, biogeographic patterns (vicariance-dispersal events) within Harpyionycterinae appear clear and can be directly linked to major biogeographic boundaries of the Austromalayan region. The new phylogeny of Harpionycterinae also provides a new framework for interpreting aspects of dental evolution in pteropodids (e.g., reduction in the incisor dentition) and allows prediction of roosting habits for Harpyionycteris, whose habits are unknown. -
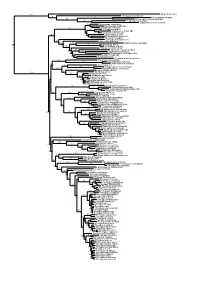
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

Avian Diversification in the Afrotropics Ben D
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 2008 Avian diversification in the afrotropics Ben D. Marks Louisiana State University and Agricultural and Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Recommended Citation Marks, Ben D., "Avian diversification in the afrotropics" (2008). LSU Doctoral Dissertations. 3021. https://digitalcommons.lsu.edu/gradschool_dissertations/3021 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. AVIAN DIVERSIFICATION IN THE AFROTROPICS A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Department of Biological Sciences by Ben D. Marks B. A., The Evergreen State College, 1995 M.S., Illinois State University, 2000 May 2008 ACKNOWLEDGMENTS I owe a great debt of gratitude to my advisor Frederick H. Sheldon for his patient support, lunchtime discussions, and careful editing of manuscripts during my years in graduate school. Time spent together in the forests of Borneo and on fishing trips will be some of my fondest memories of graduate school. I would also like to thank the other members of my graduate committee, J. V. Remsen, Michael E. Hellberg, and Robb T. Brumfield, for their time and efforts. This work would not have been possible without the efforts of many dedicated collectors, museum curators, and collection managers over several decades. -

2020 Special Issue
Journal Home page : www.jeb.co.in « E-mail : [email protected] Original Research Journal of Environmental Biology TM p-ISSN: 0254-8704 e-ISSN: 2394-0379 JEB CODEN: JEBIDP DOI : http://doi.org/10.22438/jeb/4(SI)/MS_1904 Plagiarism Detector Grammarly New records and present status of bat fauna in Mizoram, North-Eastern India C. Vanlalnghaka Department of Zoology, Govt. Serchhip College, Mizoram–796 181, India *Corresponding Author Email : [email protected] Paper received: 08.12.2019 Revised received: 24.06.2020 Accepted: 10.07.2020 Abstract Aim: The present study aimed to investigate the diversity of bat fauna in Mizoram and prepare a checklist for future references. This study also investigated threats and suggested recommendations for implementing conservation measures for bat fauna in Mizoram. Methodology: The present study was carried out in different parts of Mizoram between January 2012 - October 2019. Bats were trapped by using mist nets and hoop nets. Diagnostic morphological characters of bat were used for species identification. Digital camera and video camera were also used for further identification and documentation of bats. Results: During January 2012 – December 2016, eighteen bat species were identified. Recently, from January 2017 - October 2019 insectivorous bat species, Scotomanes ornatus was first time documented in Serchhip District (23.3 ºN 92.83 ºE), Mizoram. In total nineteen bat species were identified in this study, out of which ten species were first time recorded and nine species were rediscovered from the previous documentation. From the previous and present data, total of thirty-six bat Study the diversity of bat fauna and prepared checklist in species were recorded in Mizoram- nine Mizoram. -

Tree Species Composition and Stand Structure of Woody Savanna in Dahomey Gap
TROPICS Vol. 22 (2) 39-57 Issued September 1, 2013 Tree species composition and stand structure of woody savanna in Dahomey Gap Augustin T. B. Orou Matilo1, 2*, Yoshiko Iida3 and Takashi S. Kohyama3 1 Graduate School of Environmental Science, Hokkaido University, Sapporo 060-0810, Japan 2 Present address: Department of Forests and Natural Resources, Ministry of Environmental Affairs, BP 393, Cotonou, Benin 3 Faculty of Environmental Earth Science, Hokkaido University, Sapporo 060-0810, Japan * Corresponding author: [email protected] ABSTRACT The Dahomey gap is a savanna region separating the West African rainforests to two regions. We set two sites in this woody savanna, at north and south regions in purpose to examine the differences in tree community properties between sites and among canopy-closure types, and to reveal the factors affecting the differences. Two sites are ca. 225 km distant from each other and have different rainfall patterns. Six 1-ha plots in each site were studied, which cover three types of canopy-closure (closed, semi-open and open). We recorded 3,720 trees that are equal to or more than 5 cm diameter at the breast height, consisted of 70 species belonging to 34 families. Most abundant three species, Isoberlinia doka, Vitellaria paradoxa and Pericopsis laxiflora appeared in both sites. The detrended correspondence analysis (DCA) based on species abundance divided 12 plots into groups of north and south sites along the first axis, whereas canopy-closure types were not associated with DCA axes. At any given total tree height, trees in the north site had slenderer stems with narrower crowns than those in the south site.