A Phylogenetic Tree for Pteropus and Related Genera (Chiroptera: Pteropodidae) ⇑ Francisca C
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Predicted the Impacts of Climate Change and Extreme-Weather Events on the Future
bioRxiv preprint doi: https://doi.org/10.1101/2021.05.13.443960; this version posted May 14, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Predicted the impacts of climate change and extreme-weather events on the future distribution of fruit bats in Australia Vishesh L. Diengdoh1, e: [email protected], ORCID: https://orcid.org/0000- 0002-0797-9261 Stefania Ondei1 - e: [email protected] Mark Hunt1, 3 - e: [email protected] Barry W. Brook1, 2 - e: [email protected] 1School of Natural Sciences, University of Tasmania, Private Bag 55, Hobart TAS 7005 Australia 2ARC Centre of Excellence for Australian Biodiversity and Heritage, Australia 3National Centre for Future Forest Industries, Australia Corresponding Author: Vishesh L. Diengdoh Acknowledgements We thank John Clarke and Vanessa Round from Climate Change in Australia (https://www.climatechangeinaustralia.gov.au/)/ Commonwealth Scientific and Industrial Research Organisation (CSIRO) for providing the data on extreme weather events. This work was supported by the Australian Research Council [grant number FL160100101]. Conflict of Interest None. Author Contributions bioRxiv preprint doi: https://doi.org/10.1101/2021.05.13.443960; this version posted May 14, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -

A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies
www.nature.com/scientificreports OPEN Kanyawara Virus: A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies Received: 19 April 2017 Accepted: 25 May 2017 Infesting Previously Unknown Published: xx xx xxxx Pteropodid Bats in Uganda Tony L. Goldberg 1,2,3, Andrew J. Bennett1, Robert Kityo3, Jens H. Kuhn4 & Colin A. Chapman3,5 Bats are natural reservoir hosts of highly virulent pathogens such as Marburg virus, Nipah virus, and SARS coronavirus. However, little is known about the role of bat ectoparasites in transmitting and maintaining such viruses. The intricate relationship between bats and their ectoparasites suggests that ectoparasites might serve as viral vectors, but evidence to date is scant. Bat flies, in particular, are highly specialized obligate hematophagous ectoparasites that incidentally bite humans. Using next- generation sequencing, we discovered a novel ledantevirus (mononegaviral family Rhabdoviridae, genus Ledantevirus) in nycteribiid bat flies infesting pteropodid bats in western Uganda. Mitochondrial DNA analyses revealed that both the bat flies and their bat hosts belong to putative new species. The coding-complete genome of the new virus, named Kanyawara virus (KYAV), is only distantly related to that of its closest known relative, Mount Elgon bat virus, and was found at high titers in bat flies but not in blood or on mucosal surfaces of host bats. Viral genome analysis indicates unusually low CpG dinucleotide depletion in KYAV compared to other ledanteviruses and rhabdovirus groups, with KYAV displaying values similar to rhabdoviruses of arthropods. Our findings highlight the possibility of a yet- to-be-discovered diversity of potentially pathogenic viruses in bat ectoparasites. Bats (order Chiroptera) represent the second largest order of mammals after rodents (order Rodentia). -

Daytime Behaviour of the Grey-Headed Flying Fox Pteropus Poliocephalus Temminck (Pteropodidae: Megachiroptera) at an Autumn/Winter Roost
DAYTIME BEHAVIOUR OF THE GREY-HEADED FLYING FOX PTEROPUS POLIOCEPHALUS TEMMINCK (PTEROPODIDAE: MEGACHIROPTERA) AT AN AUTUMN/WINTER ROOST K.A. CONNELL, U. MUNRO AND F.R. TORPY Connell KA, Munro U and Torpy FR, 2006. Daytime behaviour of the grey-headed flying fox Pteropus poliocephalus Temminck (Pteropodidae: Megachiroptera) at an autumn/winter roost. Australian Mammalogy 28: 7-14. The grey-headed flying fox (Pteropus poliocephalus Temminck) is a threatened large fruit bat endemic to Australia. It roosts in large colonies in rainforest patches, mangroves, open forest, riparian woodland and, as native habitat is reduced, increasingly in vegetation within urban environments. The general biology, ecology and behaviour of this bat remain largely unknown, which makes it difficult to effectively monitor, protect and manage this species. The current study provides baseline information on the daytime behaviour of P. poliocephalus in an autumn/winter roost in urban Sydney, Australia, between April and August 2003. The most common daytime behaviours expressed by the flying foxes were sleeping (most common), grooming, mating/courtship, and wing spreading (least common). Behaviours differed significantly between times of day and seasons (autumn and winter). Active behaviours (i.e., grooming, mating/courtship, wing spreading) occurred mainly in the morning, while sleeping predominated in the afternoon. Mating/courtship and wing spreading were significantly higher in April (reproductive period) than in winter (non-reproductive period). Grooming was the only behaviour that showed no significant variation between sample periods. These results provide important baseline data for future comparative studies on the behaviours of flying foxes from urban and ‘natural’ camps, and the development of management strategies for this species. -

Checklist of the Mammals of Indonesia
CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation i ii CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation By Ibnu Maryanto Maharadatunkamsi Anang Setiawan Achmadi Sigit Wiantoro Eko Sulistyadi Masaaki Yoneda Agustinus Suyanto Jito Sugardjito RESEARCH CENTER FOR BIOLOGY INDONESIAN INSTITUTE OF SCIENCES (LIPI) iii © 2019 RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI) Cataloging in Publication Data. CHECKLIST OF THE MAMMALS OF INDONESIA: Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation/ Ibnu Maryanto, Maharadatunkamsi, Anang Setiawan Achmadi, Sigit Wiantoro, Eko Sulistyadi, Masaaki Yoneda, Agustinus Suyanto, & Jito Sugardjito. ix+ 66 pp; 21 x 29,7 cm ISBN: 978-979-579-108-9 1. Checklist of mammals 2. Indonesia Cover Desain : Eko Harsono Photo : I. Maryanto Third Edition : December 2019 Published by: RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI). Jl Raya Jakarta-Bogor, Km 46, Cibinong, Bogor, Jawa Barat 16911 Telp: 021-87907604/87907636; Fax: 021-87907612 Email: [email protected] . iv PREFACE TO THIRD EDITION This book is a third edition of checklist of the Mammals of Indonesia. The new edition provides remarkable information in several ways compare to the first and second editions, the remarks column contain the abbreviation of the specific island distributions, synonym and specific location. Thus, in this edition we are also corrected the distribution of some species including some new additional species in accordance with the discovery of new species in Indonesia. -

Zoologische Mededelingen
ZOOLOGISCHE MEDEDELINGEN UITGEGEVEN DOOR HET RIJKSMUSEUM VAN NATUURLIJKE HISTORIE TE LEIDEN (MINISTERIE VAN CULTUUR, RECREATIE EN MAATSCHAPPELIJK WERK) Deel 55 no. 14 4 maart 1980 A NEW FRUIT BAT OF THE GENUS MYONYCTERIS MATSCHIE, 1899, FROM EASTERN KENYA AND TANZANIA (MAMMALIA, MEGACHIROPTERA) by W. BERGMANS Instituut voor Taxonomische Zoölogie, Universiteit van Amsterdam With 4 text-figures ABSTRACT Myonycteris relicta n. sp. is described from the Shimba Hills in southeast Kenya and from the Usambara Mountains in northeast Tanzania. The species is larger than the only other known African mainland species of the genus, Myonycteris torquata (Dobson, 1878), from the Central and West African rain forests and, if compared to M. torquata and the only other species in the genus, M. brachycephala (Bocage, 1889) from São Tomé, has a relatively longer rostrum, a more deflected cranial axis, and further differs in number, shape and position of its teeth. The new species provides new arguments for the relationship between the genera Myonycteris Matschie, 1899, and Lissonycteris Andersen, 1912. It is believed that Myonycteris relicta may be a forest species and as such restricted to isolated East African forests. INTRODUCTION During a visit to the Zoologisches Museum in Berlin (ZMB), in April 1979, the author found two fruit bat specimens from the Tanzanian Usa- mbara Mountains, which proved to represent an undescribed taxon. Later, in June 1979, Dr C. Smeenk of the Rijksmuseum van Natuurlijke Historie at Leiden (RMNH) recognized a third specimen of this taxon in newly acquired material from the Shimba Hills in southeast Kenya. The bats differ on specific level from all other known fruit bats, and are described in the present paper. -

Chiroptera: Pteropodidae)
Chapter 6 Phylogenetic Relationships of Harpyionycterine Megabats (Chiroptera: Pteropodidae) NORBERTO P. GIANNINI1,2, FRANCISCA CUNHA ALMEIDA1,3, AND NANCY B. SIMMONS1 ABSTRACT After almost 70 years of stability following publication of Andersen’s (1912) monograph on the group, the systematics of megachiropteran bats (Chiroptera: Pteropodidae) was thrown into flux with the advent of molecular phylogenetics in the 1980s—a state where it has remained ever since. One particularly problematic group has been the Austromalayan Harpyionycterinae, currently thought to include Dobsonia and Harpyionycteris, and probably also Aproteles.Inthis contribution we revisit the systematics of harpyionycterines. We examine historical hypotheses of relationships including the suggestion by O. Thomas (1896) that the rousettine Boneia bidens may be related to Harpyionycteris, and report the results of a series of phylogenetic analyses based on new as well as previously published sequence data from the genes RAG1, RAG2, vWF, c-mos, cytb, 12S, tVal, 16S,andND2. Despite a striking lack of morphological synapomorphies, results of our combined analyses indicate that Boneia groups with Aproteles, Dobsonia, and Harpyionycteris in a well-supported, expanded Harpyionycterinae. While monophyly of this group is well supported, topological changes within this clade across analyses of different data partitions indicate conflicting phylogenetic signals in the mitochondrial partition. The position of the harpyionycterine clade within the megachiropteran tree remains somewhat uncertain. Nevertheless, biogeographic patterns (vicariance-dispersal events) within Harpyionycterinae appear clear and can be directly linked to major biogeographic boundaries of the Austromalayan region. The new phylogeny of Harpionycterinae also provides a new framework for interpreting aspects of dental evolution in pteropodids (e.g., reduction in the incisor dentition) and allows prediction of roosting habits for Harpyionycteris, whose habits are unknown. -
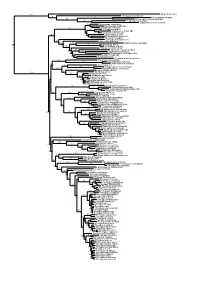
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

Priority-Setting for Philippine Bats Using Practical Approach to Guide Effective Species Conservation and Policy-Making in the Anthropocene
Published by Associazione Teriologica Italiana Volume 30 (1): 74–83, 2019 Hystrix, the Italian Journal of Mammalogy Available online at: http://www.italian-journal-of-mammalogy.it doi:10.4404/hystrix–00172-2019 Research Article Priority-setting for Philippine bats using practical approach to guide effective species conservation and policy-making in the Anthropocene Krizler Cejuela Tanalgo1,2,3,4,∗, Alice Catherine Hughes1,3 1Landscape Ecology Group, Centre for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun, Mengla, Yunnan Province 666303, People’s Republic of China 2International College, University of Chinese Academy of Sciences, Beijing 100049, People’s Republic of China 3Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Menglun, Mengla, Yunnan Province 666303, People’s Republic of China 4Department of Biological Sciences, College of Arts and Sciences, University of Southern Mindanao, Kabacan 9407, North Cotabato, the Republic of the Philippines Keywords: Abstract asian tropics endemism National level approaches to the development and implementation of effective conservation policy forest loss and practice are often challenged by limited capacity and resources. Developing relevant and hunting achievable priorities at the national level is a crucial step for effective conservation. The Philip- islands pine archipelago includes over 7000 islands and is one of only two countries considered both a oil palm global biodiversity hotspot and a megadiversity country. Yet, few studies have conducted over- arching synthesis for threats and conservation priorities of any species group. As bats make up a Article history: significant proportion of mammalian diversity in the Philippines and fulfil vital roles to maintain Received: 01/19/2019 ecosystem health and services we focus on assessing the threats and priorities to their conserva- Accepted: 24/06/2019 tion across the Philippines. -

Bat Count 2003
BAT COUNT 2003 Working to promote the long term, sustainable conservation of globally threatened flying foxes in the Philippines, by developing baseline population information, increasing public awareness, and training students and protected area managers in field monitoring techniques. 1 A Terminal Report Submitted by Tammy Mildenstein1, Apolinario B. Cariño2, and Samuel Stier1 1Fish and Wildlife Biology, University of Montana, USA 2Silliman University and Mt. Talinis – Twin Lakes Federation of People’s Organizations, Diputado Extension, Sibulan, Negros Oriental, Philippines Photo by: Juan Pablo Moreiras 2 EXECUTIVE SUMMARY Large flying foxes in insular Southeast Asia are the most threatened of the Old World fruit bats due to deforestation, unregulated hunting, and little conservation commitment from local governments. Despite the fact they are globally endangered and play essential ecological roles in forest regeneration as seed dispersers and pollinators, there have been only a few studies on these bats that provide information useful to their conservation management. Our project aims to promote the conservation of large flying foxes in the Philippines by providing protected area managers with the training and the baseline information necessary to design and implement a long-term management plan for flying foxes. We focused our efforts on the globally endangered Philippine endemics, Acerodon jubatus and Acerodon leucotis, and the bats that commonly roost with them, Pteropus hypomelanus, P. vampyrus lanensis, and P. pumilus which are thought to be declining in the Philippines. Local participation is an integral part of our project. We conducted the first national training workshop on flying fox population counts and conservation at the Subic Bay area. -

ANTIQUITY 2008 (In Press) an Unexpected, Stripe-Faced Flying Fox in Ice Age Rock Art of Australia's Kimberley. “Jack” Pett
ANTIQUITY 2008 (in press) An Unexpected, Stripe-faced Flying Fox in Ice Age Rock Art of Australia’s Kimberley. “Jack” Pettigrew, Marilyn Nugent, Anscar McPhee, Josh Wallman Bradshaw rock art of northern Australia enjoys continuing controversy concerning what community painted them and how to interpret the images (Roy 2002). The keen observation and accurate depictions of the natural world shown here, as well as the extraordinary longevity of the stains used, all tend to support the side of the controversy that posits a distinct cultural entity. We describe a painting unmistakably depicting flying foxes with features not found in bats presently found in Australia. Thermoluminescence dating of wasp nest overlying the art suggests an ice age migration to Australia, either of the bats or of the artists who painted them, a more likely scenario biologically than younger dates. The bat depictions were found on a sandstone wall protected by overhangs, near Kalumburu (14.30 °S; 126.64 °E), amongst other walls showing characteristic Bradshaw art (Walsh 2002). The depiction shows eight roosting megabats (flying foxes, Family Pteropodidae, sub-Order Megachiroptera) hanging from a slender branch, or more likely, a vine (Figs 1,2). Each bat has a distinctive white facial stripe and pale belly (Fig. 3). Figure1. White-striped flying foxes depicted in Bradshaw rock art at Kalumburu, in the Kimberley of Western Australia. Figure 2. Extension, to the left, of the same depicted group of shown in Fig. 1, with which there is some overlap. Dating: The indelible inks used in Bradshaw art penetrate more than a millimetre into the sandstone but have resisted all attempts so far to date them directly, with a number of different estimates of their age (Michaelson and Ebersole 2000). -

The Philippine Flying Foxes, Acerodon Jubatus and Pteropus Vampyrus Lanensis
Journal of Mammalogy, 86(4):719- 728, 2005 DIETARY HABITS OF THE WORLD’S LARGEST BATS: THE PHILIPPINE FLYING FOXES, ACERODON JUBATUS AND PTEROPUS VAMPYRUS LANENSIS Sam C. Stier* and Tammy L. M ildenstein College of Forestry and Conservation, University of Montana, Missoula, MT 59802, USA The endemic and endangered golden- crowned flying fox (Acerodon jubatus) coroosts with the much more common and widespread giant Philippine fmit bat (Pteropus vampyrus ianensis) in lowland dipterocarp forests throughout the Philippine Islands. The number of these mixed roost- colonies and the populations of flying foxes in them have declined dramatically in the last century. We used fecal analysis, interviews of bat hunters, and personal observations to describe the dietary habits of both bat species at one of the largest mixed roosts remaining, near Subic Bay, west- central Luzon. Dietary items were deemed “important” if used consistently on a seasonal basis or throughout the year, ubiquitously throughout the population, and if they were of clear nutritional value. Of the 771 droppings examined over a 2.5 -year period (1998-2000), seeds from Ficus were predominant in the droppings of both species and met these criteria, particularly hemiepiphytic species (41% of droppings of A. jubatus) and Ficus variegata (34% of droppings of P. v. ianensis and 22% of droppings of A. jubatus). Information from bat hunter interviews expanded our knowledge of the dietary habits of both bat species, and corroborated the fecal analyses and personal observations. Results from this study suggest that A. jubatus is a forest obligate, foraging on fruits and leaves from plant species restricted to lowland, mature natural forests, particularly using a small subset of hemiepiphytic and other Ficus species throughout the year. -

Emergence and Returning Activity in the Indian Flying Fox, Pteropus Giganteus (Chiroptera: Pteropodidae)
International Journal of Geography and Geology 1(1):1-9 International Journal of Geography and Geology journal homepage: http://aessweb.com/journal-detail.php?id=5011 EMERGENCE AND RETURNING ACTIVITY IN THE INDIAN FLYING FOX, PTEROPUS GIGANTEUS (CHIROPTERA: PTEROPODIDAE) M. R. Sudhakaran1 D. P. Swamidoss2 P. Parvathiraj3 ABSTRACT Approach: After a diurnal resting in the roost, bats adapts some behavioural pattern to get themselves active towards foraging there by involving in various activities. Their activity pattern differs from time to time depending on the change in climatic factors. But the behavioural activities they involved varies from time to time. Observation was done on the emergence and returning gate i. e the emergence or returning of first to last bat, pre emergence behaviour and post return behaviour, and influence of moonlight on foraging activity. Key Words: Post return activity, emergence gate, behaviour, pre-emergence behaviour. INTRODUCTION Most of the megachiropteran’s found in the sub tropics roost in tents, in manmade structures like old houses and temples and in foliages or hollows in trees, mainly to evade from the climatic factors, which affect them. Indian flying fox, Pteropus giganteus roosts in open foliages of trees, a peculiar character of this genus. Papers dealing with behavioural aspects of P. giganteus are scarce. Apart from Neuweiler (1969), other works on behavioural aspects focused on copulatory behaviour (Koilraj et al., 2001), roost preference (Acharya, 1936), mating (Bhatt, 1942), local migration (Breadow, 1931; Nelson, 1965a) and general ecology and biology (Brosset, 1962). Bats roosting in closed environments involved in various behavioural activities during their emergence (Kunz, 1982) like light sampling, flying inside its roost etc.